Ames test
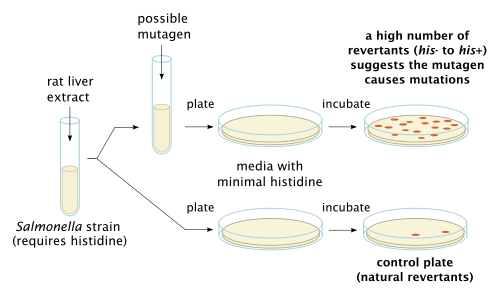
The Ames test is a widely employed method that uses bacteria to test whether a given chemical can cause mutations in the DNA of the test organism. More formally, it is a biological assay to assess the mutagenic potential of chemical compounds.[1] A positive test indicates that the chemical is mutagenic and therefore may act as a carcinogen, because cancer is often linked to mutation. The test serves as a quick and convenient assay to estimate the carcinogenic potential of a compound because standard carcinogen assays on mice and rats are time-consuming (taking two to three years to complete) and expensive. However, false-positives and false-negatives are known.[2]
The procedure was described in a series of papers in the early 1970s by Bruce Ames and his group at the University of California, Berkeley.[3][4][5][6]
General procedure
[edit]The Ames test uses several strains of the bacterium Salmonella typhimurium that carry mutations in genes involved in histidine synthesis. These strains are auxotrophic mutants, i.e. they require histidine for growth, but cannot produce it. The method tests the capability of the tested substance in creating mutations that result in a return to a "prototrophic" state, so that the cells can grow on a histidine-free medium.
The tester strains are specially constructed to detect either frameshift (e.g. strains TA-1537 and TA-1538) or point (e.g. strain TA-1531) mutations in the genes required to synthesize histidine, so that mutagens acting via different mechanisms may be identified. Some compounds are quite specific, causing reversions in just one or two strains.[4] The tester strains also carry mutations in the genes responsible for lipopolysaccharide synthesis, making the cell wall of the bacteria more permeable,[5] and in the excision repair system to make the test more sensitive.[6]
Larger organisms like mammals have metabolic processes that could potentially turn a chemical considered not mutagenic into one that is or one that is considered mutagenic into one that is not.[7] Therefore, to more effectively test a chemical compound's mutagenicity in relation to larger organisms, rat liver enzymes can be added in an attempt to replicate the metabolic processes' effect on the compound being tested in the Ames Test. Rat liver extract is optionally added to simulate the effect of metabolism, as some compounds, like benzo[a]pyrene, are not mutagenic themselves but their metabolic products are.[3]
The bacteria are spread on an agar plate with small amount of histidine. This small amount of histidine in the growth medium allows the bacteria to grow for an initial time and have the opportunity to mutate. When the histidine is depleted only bacteria that have mutated to gain the ability to produce its own histidine will survive. The plate is incubated for 48 hours. The mutagenicity of a substance is proportional to the number of colonies observed.
Ames test and carcinogens
[edit]Mutagens identified via Ames test are also possible carcinogens, and early studies by Ames showed that 90% of known carcinogens may be identified via this test.[8] Later studies however showed identification of 50–70% of known carcinogens.[citation needed] The test was used to identify a number of compounds previously used in commercial products as potential carcinogens.[9] Examples include tris(2,3-dibromopropyl)phosphate, which was used as a flame retardant in plastic and textiles such as children's sleepwear,[10] and furylfuramide which was used as an antibacterial additive in food in Japan in the 1960s and 1970s. Furylfuramide in fact had previously passed animal tests, but more vigorous tests after its identification in the Ames test showed it to be carcinogenic.[11] Their positive tests resulted in those chemicals being withdrawn from use in consumer products.
One interesting result from the Ames test is that the dose response curve using varying concentrations of the chemical is almost always linear,[8] indicating that there is no threshold concentration for mutagenesis. It therefore suggests that, as with radiation, there may be no safe threshold for chemical mutagens or carcinogens.[12][13] However, some have proposed that organisms could tolerate low levels of mutagens due to protective mechanisms such as DNA repair, and thus a threshold may exist for certain chemical mutagens.[14] Bruce Ames himself argued against linear dose-response extrapolation from the high dose used in carcinogenesis tests in animal systems to the lower dose of chemicals normally encountered in human exposure, as the results may be false positives due to mitogenic response caused by the artificially high dose of chemicals used in such tests.[15][16] He also cautioned against the "hysteria over tiny traces of chemicals that may or may not cause cancer", that "completely drives out the major risks you should be aware of".[17]
The Ames test is often used as one of the initial screens for potential drugs to weed out possible carcinogens, and it is one of the eight tests required under the Pesticide Act (USA) and one of the six tests required under the Toxic Substances Control Act (USA).[18]
Limitations
[edit]Salmonella typhimurium is a prokaryote, therefore it is not a perfect model for humans. Rat liver S9 fraction is used to mimic the mammalian metabolic conditions so that the mutagenic potential of metabolites formed by a parent molecule in the hepatic system can be assessed; however, there are differences in metabolism between humans and rats that can affect the mutagenicity of the chemicals being tested.[19] The test may therefore be improved by the use of human liver S9 fraction; its use was previously limited by its availability, but it is now available commercially and therefore may be more feasible.[20] An adapted in vitro model has been made for eukaryotic cells, for example yeast.
Mutagens identified in the Ames test need not necessarily be carcinogenic, and further tests are required for any potential carcinogen identified in the test. Drugs that contain the nitrate moiety sometimes come back positive for Ames when they are indeed safe. The nitrate compounds may generate nitric oxide, an important signal molecule that can give a false positive. Nitroglycerin is an example that gives a positive Ames yet is still used in treatment today. Nitrates in food however may be reduced by bacterial action to nitrites which are known to generate carcinogens by reacting with amines and amides. Long toxicology and outcome studies are needed with such compounds to disprove a positive Ames test.
Fluctuation method
[edit]

The Ames test was initially developed using agar plates (the plate incorporation technique), as described above. Since that time, an alternative to performing the Ames test has been developed, which is known as the "fluctuation method". This technique is the same in concept as the agar-based method, with bacteria being added to a reaction mixture with a small amount of histidine, which allows the bacteria to grow and mutate, returning to synthesize their own histidine. By including a pH indicator, the frequency of mutation is counted in microplates as the number of wells which have changed color (caused by a drop in pH due to metabolic processes of reproducing bacteria). As with the traditional Ames test, the sample is compared to the natural background rate of reverse mutation in order to establish the genotoxicity of a substance. The fluctuation method is performed entirely in liquid culture and is scored by counting the number of wells that turn yellow from purple in 96-well or 384-well microplates.
In the 96-well plate method the frequency of mutation is counted as the number of wells out of 96 which have changed color. The plates are incubated for up to five days, with mutated (yellow) colonies being counted each day and compared to the background rate of reverse mutation using established tables of significance to determine the significant differences between the background rate of mutation and that for the tested samples.
In the more scaled-down 384-well plate microfluctuation method the frequency of mutation is counted as the number of wells out of 48 which have changed color after 2 days of incubation. A test sample is assayed across 6 dose levels with concurrent zero-dose (background) and positive controls which all fit into one 384-well plate. The assay is performed in triplicates to provide statistical robustness. It uses the recommended OECD Guideline 471 tester strains (histidine auxotrophs and tryptophan auxotrophs).
The fluctuation method is comparable to the traditional pour plate method in terms of sensitivity and accuracy, however, it does have a number of advantages: it needs less test sample, it has a simple colorimetric endpoint, counting the number of positive wells out of possible 96 or 48 wells is much less time-consuming than counting individual colonies on an agar plate. Several commercial kits are available. Most kits have consumable components in a ready-to-use state, including lyophilized bacteria, and tests can be performed using multichannel pipettes. The fluctuation method also allows for testing higher volumes of aqueous samples (up to 75% v/v), increasing the sensitivity and extending its application to low-level environmental mutagens.[21]
References
[edit]- ^ Mortelmans K, Zeiger E (November 2000). "The Ames Salmonella/microsome mutagenicity assay". Mutation Research. 455 (1–2): 29–60. doi:10.1016/S0027-5107(00)00064-6. PMID 11113466.
- ^ Charnley G (2002). "Ames Test". Encyclopedia of Public Health. eNotes.com. Archived from the original on 4 February 2009. Retrieved 2014-05-02.
- ^ a b Ames BN, Durston WE, Yamasaki E, Lee FD (August 1973). "Carcinogens are mutagens: a simple test system combining liver homogenates for activation and bacteria for detection". Proceedings of the National Academy of Sciences of the United States of America. 70 (8): 2281–5. Bibcode:1973PNAS...70.2281A. doi:10.1073/pnas.70.8.2281. PMC 433718. PMID 4151811.
- ^ a b Ames BN, Gurney EG, Miller JA, Bartsch H (November 1972). "Carcinogens as frameshift mutagens: metabolites and derivatives of 2-acetylaminofluorene and other aromatic amine carcinogens". Proceedings of the National Academy of Sciences of the United States of America. 69 (11): 3128–32. Bibcode:1972PNAS...69.3128A. doi:10.1073/pnas.69.11.3128. PMC 389719. PMID 4564203.
- ^ a b Ames BN, Lee FD, Durston WE (March 1973). "An improved bacterial test system for the detection and classification of mutagens and carcinogens". Proceedings of the National Academy of Sciences of the United States of America. 70 (3): 782–6. Bibcode:1973PNAS...70..782A. doi:10.1073/pnas.70.3.782. PMC 433358. PMID 4577135.
- ^ a b McCann J, Spingarn NE, Kobori J, Ames BN (March 1975). "Detection of carcinogens as mutagens: bacterial tester strains with R factor plasmids". Proceedings of the National Academy of Sciences of the United States of America. 72 (3): 979–83. Bibcode:1975PNAS...72..979M. doi:10.1073/pnas.72.3.979. PMC 432447. PMID 165497.
- ^ Hartwell L, Goldberg M, Hood L, Reynolds A, Silver L (2011). Genetics: from genes to genomes (4th ed.). New York: McGraw-Hill. ISBN 978-0-07-352526-6. OCLC 317623365.
- ^ a b McCann J, Choi E, Yamasaki E, Ames BN (December 1975). "Detection of carcinogens as mutagens in the Salmonella/microsome test: assay of 300 chemicals". Proceedings of the National Academy of Sciences of the United States of America. 72 (12): 5135–9. Bibcode:1975PNAS...72.5135M. doi:10.1073/pnas.72.12.5135. PMC 388891. PMID 1061098.
- ^ Ames BN (May 1979). "Identifying environmental chemicals causing mutations and cancer" (PDF). Science. 204 (4393): 587–93. Bibcode:1979Sci...204..587A. doi:10.1126/science.373122. JSTOR 1748159. PMID 373122.
- ^ Prival MJ, McCoy EC, Gutter B, Rosendranz HS (January 1977). "Tris(2,3-dibromopropyl) phosphate: mutagenicity of a widely used flame retardant". Science. 195 (4273): 76–8. Bibcode:1977Sci...195...76P. doi:10.1126/science.318761. PMID 318761.
- ^ Hayatsu, Hiroka (1991), Mutagens in Food: Detection and Prevention, CRC Press, pp. 286 pages, ISBN 978-0-8493-5877-7
- ^ Teasdale A (2011). Genotoxic Impurities: Strategies for Identification and Control. Wiley-Blackwell. ISBN 978-0-470-49919-1.
- ^ Tubiana M (September 1992). "The carcinogenic effect of exposure to low doses of carcinogens". British Journal of Industrial Medicine. 49 (9): 601–5. doi:10.1136/oem.49.9.601. PMC 1039303. PMID 1390264.
- ^ Jenkins GJ, Doak SH, Johnson GE, Quick E, Waters EM, Parry JM (November 2005). "Do dose response thresholds exist for genotoxic alkylating agents?". Mutagenesis. 20 (6): 389–98. doi:10.1093/mutage/gei054. PMID 16135536.
- ^ Forman D (August 1991). "Ames, the Ames test, and the causes of cancer". BMJ. 303 (6800): 428–9. doi:10.1136/bmj.303.6800.428. PMC 1670593. PMID 1912830.
- ^ Ames BN, Gold LS (October 1990). "Chemical carcinogenesis: too many rodent carcinogens". Proceedings of the National Academy of Sciences of the United States of America. 87 (19): 7772–6. Bibcode:1990PNAS...87.7772A. doi:10.1073/pnas.87.19.7772. PMC 54830. PMID 2217209.
- ^ Twombly R (September 2001). "Federal carcinogen report debuts new list of nominees". Journal of the National Cancer Institute. 93 (18): 1372. doi:10.1093/jnci/93.18.1372. PMID 11562386.
- ^ Farmer PB, Walker JM (2006). The Molecular Basis of Cancer. Krieger Publishing Company. ISBN 978-0-7099-1044-2.
- ^ Hakura A, Suzuki S, Satoh T (January 1999). "Advantage of the use of human liver S9 in the Ames test". Mutation Research. 438 (1): 29–36. doi:10.1016/s1383-5718(98)00159-4. PMID 9858674.
- ^ Hakura A, Suzuki S, Satoh T (2004). "Improvement of the Ames test using human liver S9 preparation". In Yan Z, Caldwell G (eds.). Optimization in Drug Discovery: in vitro methods. Methods in Pharmacology and toxicology. Humana Press. ISBN 978-1-58829-332-9.
- ^ Bridges BA (November 1980). "The fluctuation test". Archives of Toxicology. 46 (1–2): 41–4. doi:10.1007/BF00361244. PMID 7235997. S2CID 23769437.
Further reading
[edit]- Phillipson, Caroline E.; Ioannides, Costas (1989-03-01). "Metabolic action of polycyclic aromatic hydrocarbons to mutagens in the Ames test by various animal species including man". Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis. 211 (1): 147–151. doi:10.1016/0027-5107(89)90115-2. ISSN 0027-5107. PMID 2493576.
- McKinnell RG (2015-11-06). The Understanding, Prevention and Control of Human Cancer: The Historic Work and Lives of Elizabeth Cavert Miller and James A. Miller. BRILL. ISBN 9789004286801.
- Claxton LD, Umbuzeiro GD, DeMarini DM (November 2010). "The Salmonella mutagenicity assay: the stethoscope of genetic toxicology for the 21st century". Environmental Health Perspectives. 118 (11): 1515–22. doi:10.1289/ehp.1002336. PMC 2974687. PMID 20682480.
