Cyclic nucleotide–gated ion channel
Cyclic nucleotide–gated ion channels or CNG channels are ion channels that function in response to the binding of cyclic nucleotides. CNG channels are nonselective cation channels that are found in the membranes of various tissue and cell types, and are significant in sensory transduction as well as cellular development. Their function can be the result of a combination of the binding of cyclic nucleotides (cGMP and cAMP) and either a depolarization or a hyperpolarization event. Initially discovered in the cells that make up the retina of the eye, CNG channels have been found in many different cell types across both the animal and the plant kingdoms. CNG channels have a very complex structure with various subunits and domains that play a critical role in their function. CNG channels are significant in the function of various sensory pathways including vision and olfaction, as well as in other key cellular functions such as hormone release and chemotaxis. CNG channels have also been found to exist in prokaryotes, including many spirochaeta, though their precise role in bacterial physiology remains unknown.
Discovery
[edit]The discovery of CNG channels is related to the discovery of intracellular messengers responsible for the mediation of responses in retinal photoreceptors. Before their discovery, it was thought that cyclic nucleotides played a role in phosphorylation. In 1985, it was discovered that cGMP was able to directly activate the light-dependent response of rod ion channels by studying light-adapted retina of frogs.[1] CNG channels were also found in cone photoreceptors, chemo sensitive cilia of olfactory sensory neurons, and the pineal gland. After the identification of amino acids from purified proteins, cloning and functional expression of CNG channels were performed. Molecular cloning allowed for the discovery of similar channels in many other tissues.[2][3] In 2000, scientists performed studies using mouse retina and molecular cloning to find a new subunit of the channel, CNG6.[4]
Function
[edit]CNG channels have important functions in signal transduction in retinal photoreceptors and olfactory receptor neurons. They are directly activated by cyclic nucleotides, and approximately 4 cyclic nucleotides are needed to activate each channel. CNG channels are nonselective and allow many alkali ions to flow into or out of a cell expressing CNG channels on its membrane. This flow of ions can result in either depolarization or hyperpolarization. CNG channels can be activated by cAMP or cGMP exclusively, or sometimes by a combination of both cNMPs, and some channels are more selective than others. Even though the activity of these channels show little voltage dependence, they are still considered voltage-dependent channels. Calcium, calmodulin, and phosphorylation modulate the opening of CNG channels.[3]
The main role of CNG channels is sensory transduction in various tissues. Many studies have shown CNG channels in rod and cone photoreceptors, and they have also been found in the brain, the heart, the kidneys, and gonads.[3]
CNG channel homologs in Caenorhabditis elegans, Drosophila melanogaster, and Limulus polyphemus have unknown functions. Studies have shown homologs in C. elegans might have functions in chemosensation.[3]
Color blindness and retinal degeneration result when CNG channels have mutations. Mutations specifically in the subunits A and B result in complete and incomplete achromatopsia.[3]


Structure
[edit]A CNG channel consists of four subunits around a central pore. Each protein subunit consists 6 transmembrane segments (S1-S6), a P-loop, intracellular amino terminal region, and carboxy terminal region. The P-loop and S6 segments around the pore, which plays a role in ion conduction. There is a cyclic nucleotide binding domain (CNBD) and connection region to the S6 segment in the carboxy terminal. There is a post-CNDB region in the amino terminal.[5]
Alpha subunits
[edit]Cyclic nucleotide gated channel alpha-subunits include
- Cyclic nucleotide-gated channel alpha 1
- Cyclic nucleotide-gated channel alpha 2
- Cyclic nucleotide-gated channel alpha 3
- Cyclic nucleotide-gated channel alpha 4
Beta subunits
[edit]Cyclic nucleotide gated channel beta-subunits include:
Pore
[edit]The structure of the pore is similar to other ion channels that contain P-loops. The P-loop enters the membrane of the pore from the extracellular side and exits to the intracellular side. The P loop enters as an alpha helix and exists as an uncoiled strand. Helices that cover the inner membrane line the channel. These also form a 6 helix bundle that signifies the entrance. In order to open the pore, a conformational change must occur in the inner 6 helix bundle.[5]
Cyclic nucleotide binding domain
[edit]A Cyclic nucleotide-binding domain is an intracellular domain located in the C-terminus region and has a similar sequence to other cyclic nucleotide-binding proteins. The domain is believed to be made up of a β-pleated sheet and two α-helices. The β-pleated sheet is made up of eight antiparallel strands. The α helices are named the B and C helices. A ligand initially binds to the β-pleated sheet, and through allosteric regulation causes the movement to an α-helix toward the β-pleated sheet. The α-helix is flexible in closed channels. When an α-helix of a CNGA1 subunit is in close proximity to another α-helix, they form intersubunit disulfide bonds. This occurs mainly in closed channels, inhibiting movement of the α-helix towards the β-pleated sheet. When a ligand binds to the β-pleated sheet, this bound cyclic nucleotide stabilizes the movement of the α-helix toward the β-pleated sheet in each subunit, pulling the α-helices away from each other.[5] [6]
C-linker
[edit]The C-linker is a region that connects the CNBD to the S6 segment. The C-linker region contributes to the contact between channel subunits as well as promotes tetramerization, the forming of tetramers. There are many residues that play a role in modulation of CNG channels. This process uses metals such as nickel, zinc, copper, and magnesium. The C-linker region is involved in the coupling of ligand binding to the opening of the pore. The C linker region forms disulfide bonds with N-terminal regions. Disulfide bonds alter the channel function therefore they most likely lie close to the tertiary structure. Disulfide bonds decrease the free energy of the open state compared to the closed state. The specific cysteine residue C481 on the C-linker region is located only a few amino acids away from the binding domain. In the closed state C481 is nonreactive; C481 must undergo a conformational change so that it is accessible for the opening of the channel. Disulfide bonds form between neighboring subunits and C481. Simultaneously there is a C35 cysteine residue at the N-terminal of the C-linker region that can reach two C481 residues, making a favorable disulfide bond compared to a C481-C481 bond.[5] [6]
S6 region
[edit]Spontaneous disulfide bond formation is state-dependent, implying that the conformational change in the helix bundle is affiliated with channel gating. When the CNG ion channels are closed, the cytoplasmic ends of the S6 helices are in close proximity to each other. Small cations are able to move through an opening, which implies that the gate is beyond the helix bundle and that S6 helices are in conjunction with conformational changes in the selectivity filter.[6]
P region
[edit]The P region forms a loop, the pore loop, connecting the S5 and S6 regions, which extend to the central axis of the channel. Ionic properties are determined by the residues in the loop between S5 and S6 transmembrane segments. The P region dictates the ion selectivity of the cyclic-nucleotide gated ion channel, which also determine the pore diameter of CNG channels. The P region functions as a channel gate since it prevents ion permeation in the closed state. The pore may be hindered by small conformational changes in this region. The P region acts as an ion selectivity filter that changes structure in the open conformation. In the open state, four identical subunits contribute a single P-loop region, which forms a selectivity filter.[6]
CNG channel family
[edit]In vertebrates, the CNG channel gene family consists of six members and belong to a larger group of voltage-gated ion channels. These genes are divided based on sequence similarity into two subtypes CNGA and CNGB.[7] The CNG channels consist of and alpha subunit, known as CNGA1 and a beta subunit CNGB1. An aspect of the alpha subunit is that it is able to form a functional channel on its own, called a homotetrameric channel. This homotetrameric channel can be of scientific interest because it can be used to further understand the nature of ligand binding and selectivity of a particular channel of interest. Additional genes that code for CNG channels have been cloned from Caenorhabditis elegans and Drosophila melanogaster. A subunit of a CNG channel CNGA1, previously called the rod α subunit, was expressed in rod photoreceptors and produced functional channels that were gated by cGMP when expressed externally in either Xenopus oocytes or in a human embryonic kidney cell line (HEK293). In humans, mutated CNGA1 genes result in an autosomal recessive form of retinitis pigmentosa, a degenerative form of blindness. CNGB1, previously called the rod β subunit, is a second subunit of the rod channel. Unlike CNGA1, CNGB1 subunits expressed alone do not produce functional CNG channels, but coexpression of CNGA1 and CNGB1 subunits produces heteromeric channels with modulation, permeation, pharmacology, and cyclic-nucleotide specificity comparable to that of native channels.[8]
CNG channels form tetramers, and recent studies indicate that native rod channels consist of three CNGA1 subunits and one CNGB1 subunit. CNGA3 subunits, previously called the cone α subunits, form functional channels when its expression occurs exogenously. On the other hand, CNGB3, previously called the cone β subunit, does not. Mutations in human CNGA3 and CNGB3 are involved in complete achromatopsia, which is a rare, autosomal recessive inherited and congenital disorder characterized by the complete failure in color distinction.[8]
CNGA2, previously called the olfactory α subunit, CNGA4, previously called the olfactory β subunit, and CnGB1b are involved in transduction of odorant signals in olfactory sensory neurons for which the subunit stoichiometry and arrangement are unknown.[8]
In invertebrates, a CNG channel subunit called CNG-P1 has been cloned from D. melanogaster and is expressed in antennae and the visual system, an indication that CNG channels may be linked to the transduction of light in invertebrates. A second putative CNG-like subunit called CNGL, cloned from D. melanogaster, is found to be expressed in the brain. Two CNG channel subunits, Tax-2 and Tax-4, have been cloned in C. elegans and are responsible for chemosensation, thermosensation, and normal axon outgrowth of some sensory neurons in C. elegans.[8]
The binding event
[edit]The ligand might be placed at the bottom of the cavity due to interactions with the phosphate binding cassette (PBC). This cavity refers to a region in the CNBD formed by the β roll, a two-looped β helix. Changes induced by ligand binding occur in α helices (αA, αB, and αC and PBC helix). The β roll only undergoes small changes during binding. After the ligand is seated, αB and αC helices arrange themselves so that they form a cap over the cavity. How binding affects the αA helix is still unclear. [9]
Cooperative and non-cooperative activation
[edit]The steep concentration between CNG channels and ligand concentration shows that at least two or three cyclic nucleotides are needed. It is believed that the second ligand is required for the channel to transition from closed to open. When the third and fourth ligands bind, the open state of the channel becomes stabilized.[9] In bacteria, the opening of CNG channels is the result of non-cooperative binding.[9] With differing concentrations of ligands, cooperative binding and non-cooperative binding arise to adapt to these differing environments. At low ligand concentrations, it is rare for a ligand to cooperatively bind, because cooperative binding at low concentrations weakens the binding between channel and ligand, reducing channel sensitivity.[9]
Ligand selectivity
[edit]By measuring the currents activated in excised inside-out membrane patches upon superfusion with varying ligand concentrations, the ligand sensitivity and selectivity of both endogenously and exogenously expressed CNG channels have been studied. All native CNG channels react to both cAMP and cGMP, but smaller concentrations of cGMP than of cAMP are needed to activate and open the channels. CNG channels are sharply selective between cGMP and cAMP in rods and cones, whereas in OSNs, the channels respond equally well to both ligands. CNG channels found in OSNs are much more sensitive to both cGMP and cAMP than photoreceptor CNG channels. Studies of dose response relations showed that channel activation is greatly dependent on cGMP concentration; several cGMP molecules bind to the channel in a cooperative manner. Since each subunit contains a single cNMP-binding site, and homomeric and heteromeric channels most likely form a tetrameric complex, a maximum of four ligand molecules can bind to the channel. Selectivity can be achieved by differential control of the affinity for binding of the ligand, efficacy of gating, or a combination of both. Binding affinity means how tightly cyclic nucleotides bind to the channel. Efficacy refers to the ability of ligand to activate and open the channel once it is bound. Although these processes are useful in understanding selectivity, they are inextricably coupled to each other that it is very difficult to experimentally separate one from another.[3]
CNG channels do not discriminate between Na+ and K+ ions, and they also allow Ca2+ and Mg2+ to pass through, although at a slower rate. The passage of these divalent ions inhibit the current carried by Na+ and K+. A highly conserved residue of glutamic acid in the selectivity filter of CNG channels has been found to form a high-affinity binding site for Ca2+. Moreover, a bacterial nonselective cation channel called the NaK channel hosts a selectivity filter sequence similar to that of CNG channels. In the crystal structure of the NaK channel, a discrete Ca2+-binding site at the extracellular opening of the pore has been identified.[9]
Inhibition of CNG channels
[edit]Studies have shown the differential inhibition of CNG channels by diacylglycerol (DAG) in Xenopus oocytes. DAG can be used as a closed state inhibitor. Using DAG to inhibit homomultimeric rod channels, or those that contain two or more identical peptide chains, was similar to the inhibition of native rod channels. Using DAG to inhibit the homomultimeric olfactory channels was not as effective, even with a high concentration of DAG. More than one molecule of DAG is needed to inhibit a channel effectively. DAG molecules position themselves to stabilize the close state of a CNG channel, by binding to the channel or by changing the interaction between the lipid bilayer of the cell membrane and the channel. Studies with chimeras rod and olfactory channels suggest that differences in DAG inhibition are due to differences in the attached loop of the transmembrane segment.[10]
Physiological significance
[edit]Photoreceptors
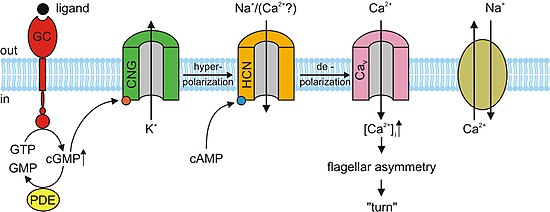
[edit]In the absence of light, cGMP binds to CNG channels in photoreceptors. This binding causes the channels to open, which allows sodium (Na+) and calcium (Ca2+) ions to flow into the cell causing the outer segment of the photoreceptor to depolarize. This depolarizing flow of ions is known as the dark current. When the retina of the eye detects light, a reaction known as a phototransduction cascade occurs. It is a signal transduction pathway that leads to the activation of the enzyme phosphodiesterase, which hydrolyzes cGMP into 5’-GMP, decreasing the concentration of cGMP. In the absence of cGMP, the CNG channels in the photoreceptors close preventing the flow of the aforementioned dark current. This in turn causes a hyperpolarization of the outer segment of the photoreceptor, preventing the propagation of an action potential and the release of glutamate.[3][5] Studies have shown that over activation of cGMP-dependent CNG channels in photoreceptors can lead to their degeneration. If the CNG channels on a photoreceptor are continuously activated, Ca2+ and Na+ ion flux into the outer segment of the photoreceptor will increase so that it depolarizes beyond the dark current. Through a positive feedback loop, this would then increase the current of Ca2+ into the cell. High concentration of Ca2+ in the photoreceptor cell would lead to its death programmed cell death or apoptosis.[11]
Retinitis pigmentosa
[edit]
Retinitis Pigmentosa (RP) is a genetic disease in which patients suffer degeneration of rod and cone photoreceptors. The loss starts in the patient's peripheral vision and progresses to the central visual field, leaving the patient blind by middle age.
About 1% of RP patients have mutations in cGMP alpha-subunit. Eight mutations have been identified- four are nonsense mutations, one is a deletion that includes most of the transcriptional unit. The other three are missense mutations and frameshift mutations, which lead to a shortening of amino acid sequence in the C terminus. It is still not known why the absence of cGMP-gated cation channels causes photoreceptor degradation. Mutations causing RP have also been found in the rhodopsin gene and in the alpha- and beta-subunits of rod phosphodiesterase, which encode rod phototransduction cascades. The mutation of these subunits indirectly impairs rod cGMP-gated channel function, which implies that there is a common mechanism of photoreceptor degradation.[12]
Pacemaker cells
[edit]In the nervous system, heart, and some visceral organs, cells contain cyclic nucleotide gated channels which determine the rhythm of the organ. These channels, formally called hyperpolarization-activated cyclic nucleotide–gated channels (HCN channels), are also termed "pacemaker channels" because of this critical function. As their name implies, they are open during conditions of hyperpolarization and closed during depolarization. The significance of this in the sinoatrial node (and, as backup, in the atrioventricular node) is that as the heart resets, or hyperpolarizes, after each beat, HCN channels open, allowing positive ions to rush into the cell (the so-called funny current), triggering another depolarization event and subsequent cardiac contraction. This gives the heart its automaticity. The primary cyclic nucleotide operating in conjunction with the HCN channel is cAMP.[13]
Olfactory sensory neurons
[edit]Almost all responses to odorants in olfactory sensory neurons (OSNs) are facilitated by CNG channels. When an odorant binds to its specific receptor in the chemosensitive cilia membrane, it activates a G protein, which causes a downstream reaction activating the enzyme adenylyl cyclase (AC). This enzyme is responsible for an increase in cAMP concentration within the OSN. cAMP binds to the CNG channels in the OSN membrane, opening them, and making the cell highly permeable to Ca2+. Calcium ions flow into the cell causing a depolarization. As in all other cell types, CNG channels in OSNs also allow Na+ to flow into the cell. Additionally, the increased Ca2+ concentration inside the cell activates Ca2+-dependent chloride (Cl−) channels, which causes intracellular Cl− ions to also flow out of the cell augmenting the depolarization event. This depolarization stimulates an action potential that ultimately signals the reception of the odorant. In addition to cAMP gated ion channels, a small subset of OSNs also has cGMP-selective CNG channels that contain the CNGA3 subunit.[3]
Spermatozoa
[edit]cAMP and cGMP mediate several cellular responses such as acrosomal exocytosis, or the fusion of the male sperm to the female egg, and chemotaxis. In the sea urchin species, Strongylocentrotus purpuratus, speract, a short peptide, was studied. Speract activates a receptor-type guanylate cyclase (GC) and stimulates a rise in intracellular cGMP concentrations. Speract also increases the concentration of calcium. Although there has yet to be any establishment of a direct causal relationship, the previously mentioned observations suggest that cGMP activates calcium conductance. CNG channels are prime candidates for the calcium-entry pathway, due to their high calcium permeability. CNG channels have yet to be detected by homology screening. In mammals, the testicular CNG channel subunits that are expressed are A3, B1, and B3. Heterologous expression of the A3 subunit was cloned from testis and produced channels that were cGMP sensitive and selective. It is possible that these channels are involved in a cGMP-stimulated calcium flux into the sperm. However a more extensive characterization of the channel has not been accomplished due to the low success rate of detecting channel activity. Since A3 subunit knockout mice are fertile, CNG channels could be involved in some form of motility control and even in chemotactic swimming behavior or in the acrosomal exocytosis. However, a receptor-type GC in mammalian sperm has yet to be identified. Mouse sperm express other channels such as CatSper1. Male sterility can be accomplished by disrupting the CatSper1 gene; additionally, the cAMP-induced calcium influx is abolished in mutant mice. Although CatSper need additional subunits to become functional, they are unrelated to CNG channels because CatSper lacks a cAMP/cGMP-binding site. It is possible that CNG and CatSper subunits assemble to form calcium-permeable and cyclic nucleotide-sensitive ion channels.[3]
Kidney
[edit]cGMP-sensitive channels have been analyzed in the renal medulla, specifically in collecting duct cells, which influence the body's electrolyte and fluid balance. CNG channel activity is controlled by the interaction between cGMP-dependent protein kinase and G1 protein because of cGMP's involvement in phosphorylation mechanisms. In the cells from an inner medullary collecting duct, CNG channels exhibit cation selectivity unit conductance, calcium permeability, and pharmacology very similar to cyclic nucleotide-gated ion channels. A stimulant, atrial natriuretic factor (ANF) increases cGMP production in the kidneys, which increases function of the glomerulus by a combination of relaxing and contracting arterioles.[14] Differences between retinal and renal cDNA have been implicated in the functional differences between CNG channels in these two tissues.[3]
Gonadotropin-releasing hormone
[edit]There has been identification of CNG ion channel subunits A2, A4, and B1 in a neuronal cell line that secretes gonadotropin-releasing hormone (GrH). The three subunits make up the CNG channels on chemosensitive cilia of OSNs. In high extracellular calcium, the unit conductance of CNG channels in rods and OSNs are significantly smaller than those measured in the neuronal line. It seems doubtful that CNG channels would create large unit conductance.[3]

Plants
[edit]CNG ions channels in plants are similar in amino acid sequence and structure to non-selective cation CNG channels in animals, as well as trans-membrane-domain K+-selective shaker family channels. However, there are drastic differences that are seen exclusively in plant CNG channels. The amino acid sequence of the pore sequence in plant CNG channels lacks the selectivity filter found in animal CNG channels as well as lacks a glycine-tyrosine-glycine-aspartate (GYGD) motif in the K+-selectivity filter sequence. Other sequence differences are seen in plant CNG channels, particularly in the cyclic nucleotide binding domain (CNBD). In plants, the Calmodulin binding domain (CaMBD) is found to overlap α-helix C in the CNBD of CNG channels. In animals the CaMBDs are located far away from the CNBD.[15] CNG channels play a large role in plant immunity and response to pathogens or external infectious agents. They have also been implicated in apoptosis in plants. CNG ion channels are also thought to be involved in pollen development in plants, however its exact role in this mechanism is still not known.[15]
Unlike animal CNG channels, plant CNG channels have not been extensively analyzed biochemically with respect to their structure.[15]
Prokaryotes
[edit]CNG ion channels share a high degree of sequence and structural similarity to mammalian CNG channels.[16] Like mammalian CNG channels, binding of cyclic nucleotides to the CNBDs has been shown to regulate channel activity and alter the channel conformational state.[16][17] Because these channels were only recently identified in spirochaeta and leptospira species,[16] their precise physiological function remains unknown in these organisms. In combination with photoactivated adenylyl cyclases, they have been used as optogenetic tools to inhibit action potential generation in neurons.[18]
Current and future research
[edit]Researchers have answered many important questions regarding CNG ion channels functions in vision and olfaction. In other physiological areas, the role of CNG channels is less defined. With technological growth, there now exists more possibilities for understanding these mechanisms.[3]
Because nitric oxide (NO) is involved in stimulating the synthesis of cGMP, further research is being conducted to understand the physiological interaction of NO with CNG channels, particularly in the covalent modification of CNG channels in OSNs.[3]
Scientists are adding on to the mechanism involved in the interaction of binding sites and interfaces of subunits. This might be nonexistent in non-cooperative CNG channels. It is also possible that binding site and gate are attached to a single subunit. In order to develop these ideas, double electron-electron resonance (DEER) and rapid fixing techniques can show these mechanistic movements.[9]
A 2007 study suggests that because of the various and complex regulatory properties in addition to the large number of CNG channels in plants, a multidisciplinary study to research plant CNG channels should be conducted.[15] Another study in March 2011 recognizes recent reverse genetics data that has been helpful in further understanding CNG channels in plants, and also suggests that additional research be conducted to identify the upstream and downstream factors in CNGC-mediated signal transduction in plants.[19]
Scientist are speculating whether DAG directly binds with CNG channel during inhibition. It is possible that DAG may insert itself into the transmembrane domains in the channel. It is also possible that DAG inserts itself into the interface between the channel and bilayer. The molecular mechanism of DAG inhibition is still not fully understood.[10]
See also
[edit]References
[edit]- ^ Fesenko EE, Kolesnikov SS, Lyubarsky AL (1985). "Induction by cyclic GMP of cationic conductance in plasma membrane of retinal rod outer segment". Nature. 313 (6000): 310–3. Bibcode:1985Natur.313..310F. doi:10.1038/313310a0. PMID 2578616. S2CID 4315094.
- ^ Yau KW (April 1994). "Cyclic nucleotide-gated channels: an expanding new family of ion channels". Proc. Natl. Acad. Sci. U.S.A. 91 (9): 3481–3. Bibcode:1994PNAS...91.3481Y. doi:10.1073/pnas.91.9.3481. PMC 43603. PMID 7513422.
- ^ a b c d e f g h i j k l m Kaupp UB, Seifert R; Seifert (July 2002). "Cyclic nucleotide-gated ion channels". Physiol. Rev. 82 (3): 769–824. CiteSeerX 10.1.1.319.7608. doi:10.1152/physrev.00008.2002. PMID 12087135.
- ^ Gerstner A, Zong X, Hofmann F, Biel M (February 2000). "Molecular Cloning and Functional Characterization of a New Modulator y Cyclic Nucleotide-Gated Channel Subunit from Mouse Retina". The Journal of Neuroscience. 20 (4): 1324–1332. doi:10.1523/JNEUROSCI.20-04-01324.2000. PMC 6772363. PMID 10662822.
- ^ a b c d e Matulef, Kimberly; Zagotta, William N. (2003). "Cyclic Nucleotide-Gated Ion Channels". Annual Review of Cell and Developmental Biology. 19 (1): 23–44. doi:10.1146/annurev.cellbio.19.110701.154854. PMID 14570562.
- ^ a b c d Wang, Zengchao; Jaing, Yongqing; Lizhi Lu; Ruihua Huang; Qingchao Hou; Fangxiong Shi (2007). "Molecular Mechanisms of Cyclic Nucleotide-Gated Ion Channel Gating". Journal of Genetics and Genomics. 34 (6): 477–485. doi:10.1016/S1673-8527(07)60052-6. PMID 17601606.
- ^ Bradley, Jonathan (2001). "Nomenclature for Ion Channel Subunits". Science. 294 (5549): 2095–2096. doi:10.1126/science.294.5549.2095. PMC 2901924. PMID 11764791.
- ^ a b c d Matulef K, Zagotta WN; Zagotta (2003). "Cyclic nucleotide-gated ion channels". Annu. Rev. Cell Dev. Biol. 19 (1): 23–44. doi:10.1146/annurev.cellbio.19.110701.154854. PMID 14570562.
- ^ a b c d e f Cukkamane A, Seifert R, Kaupp UB (January 2011). "Cooperative and uncooperative cyclic-nucleotide-gated ion channels". Trends in Biochemical Sciences. 36 (1): 55–64. doi:10.1016/j.tibs.2010.07.004. PMID 20729090.
- ^ a b Crary JI, Dean DM, Nguitragool W, Kurshan PT, Zimmerman AL (December 2000). "Mechanism of Inhibition of Cyclic Nucleotide-Gated Ion Channels by Diacylglycerol". The Journal of General Physiology. 116 (6): 755–768. doi:10.1085/jgp.116.6.755. PMC 2231817. PMID 11099345.
- ^ Vallazza-Deschamps, Géraldine; Cia, David; Gong, Jie; Jellali, Abdeljelil; Forster, Valérie; Sahel, Jose A; Tessier, Luc-Henri; Picaud, Serge (2005). "Excessive activation of cyclic nucleotide-gated channels contributes to neuronal degeneration of photoreceptors". European Journal of Neuroscience. 22 (5): 1013–1022. doi:10.1111/j.1460-9568.2005.04306.x. PMID 16176343. S2CID 36480526.
- ^ Ashcroft, Frances (1999). Ion Channels and Disease. Academic Press. ISBN 978-0-12-065310-2.
- ^ Larsson, H. P. (2010). "How is the heart rate regulated in the sinoatrial node? Another piece to the puzzle". The Journal of General Physiology. 136 (3): 237–241. doi:10.1085/jgp.201010506. PMC 2931147. PMID 20713549.
- ^ Biel, Martin; Zong, Xiangang; Distler, Madeleine; Bosse, Eva; Klugbauer, Norbert; Murakami, Manabu; Flockerzi, Veit; Hofmann, Franz (1994). "Another member of the cyclic nucleotide-gated channel family, expressed in testis, kidney, and heart". Proceedings of the National Academy of Sciences of the United States of America. 91 (9): 3505–3509. Bibcode:1994PNAS...91.3505B. doi:10.1073/pnas.91.9.3505. PMC 43608. PMID 8170936.
- ^ a b c d Kaplan B, Sherman T, Fromm H (May 2007). "Cyclic nucleotide-gated channels in plants". FEBS Lett. 581 (12): 2237–46. doi:10.1016/j.febslet.2007.02.017. PMID 17321525.
- ^ a b c Brams, M.; Kusch, J.; Spurny, R.; Benndorf, K.; Ulens, C. (2014). "Family of prokaryote cyclic nucleotide-modulated ion channels". Proceedings of the National Academy of Sciences. 111 (21): 7855–7860. Bibcode:2014PNAS..111.7855B. doi:10.1073/pnas.1401917111. ISSN 0027-8424. PMC 4040583. PMID 24821777.
- ^ James, Zachary M.; Borst, Andrew J.; Haitin, Yoni; Frenz, Brandon; DiMaio, Frank; Zagotta, William N.; Veesler, David (2017). "CryoEM structure of a prokaryotic cyclic nucleotide-gated ion channel". Proceedings of the National Academy of Sciences. 114 (17): 4430–4435. Bibcode:2017PNAS..114.4430J. doi:10.1073/pnas.1700248114. ISSN 0027-8424. PMC 5410850. PMID 28396445.
- ^ Beck, Sebastian; Yu-Strzelczyk, Jing; Pauls, Dennis; Constantin, Oana M.; Gee, Christine E.; Ehmann, Nadine; Kittel, Robert J.; Nagel, Georg; Gao, Shiqiang (2018). "Synthetic Light-Activated Ion Channels for Optogenetic Activation and Inhibition". Frontiers in Neuroscience. 12: 643. doi:10.3389/fnins.2018.00643. ISSN 1662-453X. PMC 6176052. PMID 30333716.
- ^ Moeder, Wolfgang; Urquhart, William; Ung, Huoi; Yoshioka, Keiko (2011). "The Role of Cyclic Nucleotide-Gated Ion Channels in Plant Immunity". Molecular Plant. 4 (3): 442–452. doi:10.1093/mp/ssr018. PMID 21459831.
