GNAS complex locus
GNAS complex locus is a gene locus in humans. Its main product is the heterotrimeric G-protein alpha subunit Gs-α, a key component of G protein-coupled receptor-regulated adenylyl cyclase signal transduction pathways. GNAS stands for Guanine Nucleotide binding protein, Alpha Stimulating activity polypeptide.[5]
Gene
[edit]This gene locus has a highly complex imprinted expression pattern. It gives rise to maternally-, paternally- and biallelically-expressed transcripts that are derived from four alternative promoters with distinct 5' exons. Some transcripts contain a differentially methylated region (DMR) within their 5' exons; such DMRs are commonly found in imprinted genes and correlate with transcript expression. An antisense transcript also exists, and this antisense transcript and one of the sense transcripts are paternally expressed, produce non-coding RNAs and may regulate imprinting in this region. In addition, one of the transcripts contains a second frame-shifted open reading frame, which encodes a structurally unrelated protein named ALEX.[6][7]
Products and functions
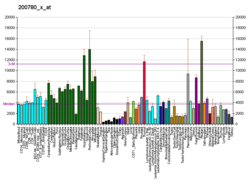
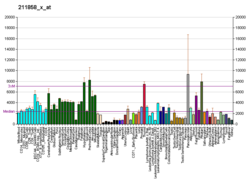
[edit]The GNAS locus is imprinted and encodes 5 main transcripts:
- Gs-α (Gs-α long, P63092-1), biallelic
- A/B transcript (Gs-α short, P63092-2), biallelic: contains an alternate 5' terminal exon (A/B or Exon 1A) and uses a downstream start codon to have a shortened amino terminal region.
- STX16 deletion causes loss of methylation at the A/B exon, leading to PHP1B.
- XLαs (Extra long alpha-s, Q5JWF2), paternal
- ALEX (Alternative gene product encoded by XL-exon, P84996), may inhibit XLαs
- NESP55 (Neuroendocrine secretory protein 55, O95467), maternal
- antisense GNAS transcript (Nespas: neuroendocrine secretory protein antisense)
Alternative splicing of downstream exons is also observed, which results in different forms of the Gs-α, a key element of the classical signal transduction pathway linking receptor-ligand interactions with the activation of adenylyl cyclase and a variety of cellular responses. Multiple transcript variants have been found for this gene, but the full-length nature and/or biological validity of some variants have not been determined.
Three of the GNAS gene products, Gsα-long, Gsα-short, and XLαs, are different forms of Gsα, and differ mainly in the N-terminal region. Traditional G protein-coupled receptor signaling proceeds primarily through Gsα-long and Gsα-short, the most abundant, ubiquitously-expressed protein products of this gene. XLαs is the "extra large" isoform, and has a very long N-terminal region with some internal repeats not well-conserved across species. The XL exon also encodes in another reading frame the protein product ALEX, an inhibitory cofactor binding to the unique domain.[10][7] The structure for GNAS is solved for the canonical P63092-1 isoform only, and little is known about what the special region of XLas or ALEX looks like.
NESP55 is a protein product completely unrelated to the GNAS protein. It undergoes extensive posttranslation processing, and is sometimes grouped as a granin.[11] Nearly nothing is known about its structure; protein structure prediction predicts a mostly disordered protein with an N-terminal globular domain made up of alpha-helices.[12][13]
Clinical significance
[edit]Mutations in GNAS products are associated with:
- Albright hereditary osteodystrophy
- pseudohypoparathyroidism type Ia and Ib
- pseudopseudohypoparathyroidism
- McCune–Albright syndrome
- Myxoma[14]
Mutations in this gene also result in progressive osseous heteroplasia, polyostotic fibrous dysplasia of bone, and some pituitary tumors.[15] Mutations in the repeat region of the XL exon leads to a hyperactive form of XLas due to lowered interaction with ALEX. As XLas is expressed in platelets, the risk of bleeding is elevated.[16][10]
Many alleles in mice have been constructed for analyzing disease associations. Mice with this gene half knocked-out and half-mutated (tm1Jop/Oedsml) display increased heart weight, increased startle reflex, and abnormalities in bone structure and mineralization;[17] some other alternations can be lethal.[18] Metabolic problems resembling pseudohypoparathyroidism are seen in heterozygous mutated (wt/Oedsml) mice.[19] Knocking out the antisense transcript is known to, at minimum, cause methylation defects.[20]
Interactions
[edit]G protein-coupled receptor-activated Gsα binds to the enzyme adenylyl cyclase, increasing its rate of conversion of ATP to cyclic AMP.[21]
Gsα has been shown to interact with RIC8A.[22]
References
[edit]- ^ a b c GRCh38: Ensembl release 89: ENSG00000087460 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000027523 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Symbol report for GNAS". HUGO Gene Nomenclature Committee.
- ^ Klemke M, Kehlenbach RH, Huttner WB (July 2001). "Two overlapping reading frames in a single exon encode interacting proteins--a novel way of gene usage". The EMBO Journal. 20 (14): 3849–60. doi:10.1093/emboj/20.14.3849. PMC 125537. PMID 11447126.
- ^ a b Abramowitz J, Grenet D, Birnbaumer M, Torres HN, Birnbaumer L (June 2004). "XLalphas, the extra-long form of the alpha-subunit of the Gs G protein, is significantly longer than suspected, and so is its companion Alex". Proceedings of the National Academy of Sciences of the United States of America. 101 (22): 8366–71. Bibcode:2004PNAS..101.8366A. doi:10.1073/pnas.0308758101. PMC 420400. PMID 15148396.
- ^ Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, Song JJ, Kingston RE, Borowsky M, Lee JT (December 2010). "Genome-wide identification of polycomb-associated RNAs by RIP-seq". Molecular Cell. 40 (6): 939–53. doi:10.1016/j.molcel.2010.12.011. PMC 3021903. PMID 21172659.
- ^ "Nespas". Long non-coding RNA db. Archived from the original on 18 June 2017. Retrieved 3 May 2019.
- ^ a b Freson K, Jaeken J, Van Helvoirt M, de Zegher F, Wittevrongel C, Thys C, Hoylaerts MF, Vermylen J, Van Geet C (May 2003). "Functional polymorphisms in the paternally expressed XLalphas and its cofactor ALEX decrease their mutual interaction and enhance receptor-mediated cAMP formation". Human Molecular Genetics. 12 (10): 1121–30. doi:10.1093/hmg/ddg130. PMID 12719376.
- ^ Bartolomucci A, Possenti R, Mahata SK, Fischer-Colbrie R, Loh YP, Salton SR (December 2011). "The extended granin family: structure, function, and biomedical implications". Endocrine Reviews. 32 (6): 755–97. doi:10.1210/er.2010-0027. PMC 3591675. PMID 21862681.
- ^ Jianwei Zhu, Sheng Wang, Dongbo Bu, Jinbo Xu. "Result for NESP55". RaptorX. Archived from the original on 4 May 2019. Retrieved 4 May 2019. Compare outputs
- ^ "O95467". MobiDB. Retrieved 4 May 2019.
- ^ Delaney D, Diss TC, Presneau N, Hing S, Berisha F, Idowu BD, O'Donnell P, Skinner JA, Tirabosco R, Flanagan AM (May 2009). "GNAS1 mutations occur more commonly than previously thought in intramuscular myxoma". Modern Pathology. 22 (5): 718–24. doi:10.1038/modpathol.2009.32. PMID 19287459.
- ^ "Entrez Gene: GNAS GNAS complex locus".
- ^ Freson K, Hoylaerts MF, Jaeken J, Eyssen M, Arnout J, Vermylen J, Van Geet C (September 2001). "Genetic variation of the extra-large stimulatory G protein alpha-subunit leads to Gs hyperfunction in platelets and is a risk factor for bleeding". Thrombosis and Haemostasis. 86 (3): 733–8. doi:10.1055/s-0037-1616126. PMID 11583302. S2CID 34153703.
- ^ "Gnas - GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus". International Mouse Phenotyping Consortium. Retrieved 3 May 2019.
- ^ "Gnas Phenotype Annotations". Mouse Genome Informatics.
- ^ "Gnas Chemically induced Allele Detail MGI Mouse (MGI:2183318)". Mouse Genome Informatics. Retrieved 3 May 2019.
- ^ "Nespas Phenotype Annotations". Mouse Genome Informatics.
- ^ Hanoune J, Defer N (April 2001). "Regulation and role of adenylyl cyclase isoforms". Annual Review of Pharmacology and Toxicology. 41 (1): 145–74. doi:10.1146/annurev.pharmtox.41.1.145. PMID 11264454.
- ^ Klattenhoff C, Montecino M, Soto X, Guzmán L, Romo X, García MA, Mellstrom B, Naranjo JR, Hinrichs MV, Olate J (May 2003). "Human brain synembryn interacts with Gsalpha and Gqalpha and is translocated to the plasma membrane in response to isoproterenol and carbachol". Journal of Cellular Physiology. 195 (2): 151–7. doi:10.1002/jcp.10300. hdl:10533/174200. PMID 12652642. S2CID 84975473.
Further reading
[edit]- Tinschert S, Gerl H, Gewies A, Jung HP, Nürnberg P (March 1999). "McCune-Albright syndrome: clinical and molecular evidence of mosaicism in an unusual giant patient". American Journal of Medical Genetics. 83 (2): 100–8. doi:10.1002/(SICI)1096-8628(19990312)83:2<100::AID-AJMG5>3.0.CO;2-K. PMID 10190480.
- Faivre L, Nivelon-Chevallier A, Kottler ML, Robinet C, Khau Van Kien P, Lorcerie B, Munnich A, Maroteaux P, Cormier-Daire V, LeMerrer M (March 2001). "Mazabraud syndrome in two patients: clinical overlap with McCune-Albright syndrome". American Journal of Medical Genetics. 99 (2): 132–6. doi:10.1002/1096-8628(2000)9999:999<00::AID-AJMG1135>3.0.CO;2-A. PMID 11241472.
- Raymond JR, Mukhin YV, Gelasco A, Turner J, Collinsworth G, Gettys TW, Grewal JS, Garnovskaya MN (2002). "Multiplicity of mechanisms of serotonin receptor signal transduction". Pharmacology & Therapeutics. 92 (2–3): 179–212. doi:10.1016/S0163-7258(01)00169-3. PMID 11916537.
- Weinstein LS, Chen M, Liu J (June 2002). "Gs(alpha) mutations and imprinting defects in human disease". Annals of the New York Academy of Sciences. 968 (1): 173–97. Bibcode:2002NYASA.968..173W. doi:10.1111/j.1749-6632.2002.tb04335.x. PMID 12119276. S2CID 85149630.
- Bastepe M, Jüppner H (2005). "GNAS locus and pseudohypoparathyroidism". Hormone Research. 63 (2): 65–74. doi:10.1159/000083895. PMID 15711092.
- de Sanctis L, Delmastro L, Russo MC, Matarazzo P, Lala R, de Sanctis C (May 2006). "Genetics of McCune-Albright syndrome". Journal of Pediatric Endocrinology & Metabolism. 19 (Suppl 2): 577–82. doi:10.1515/jpem.2006.19.s2.577. PMID 16789620. S2CID 33555734.
- Aldred MA (May 2006). "Genetics of pseudohypoparathyroidism types Ia and Ic". Journal of Pediatric Endocrinology & Metabolism. 19 (Suppl 2): 635–40. doi:10.1515/jpem.2006.19.s2.635. PMID 16789628. S2CID 26538688.
- Jüppner H, Bastepe M (May 2006). "Different mutations within or upstream of the GNAS locus cause distinct forms of pseudohypoparathyroidism". Journal of Pediatric Endocrinology & Metabolism. 19 (Suppl 2): 641–6. doi:10.1515/jpem.2006.19.s2.641. PMID 16789629. S2CID 34302323.
- Mantovani G, Spada A (December 2006). "Mutations in the Gs alpha gene causing hormone resistance". Best Practice & Research. Clinical Endocrinology & Metabolism. 20 (4): 501–13. doi:10.1016/j.beem.2006.09.001. PMID 17161328.
External links
[edit]- GNAS+protein,+human at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
- WikiGene index for literature mentioning this gene: