EGR1
EGR-1 (Early growth response protein 1) also known as ZNF268 (zinc finger protein 268) or NGFI-A (nerve growth factor-induced protein A) is a protein that in humans is encoded by the EGR1 gene.
EGR-1 is a mammalian transcription factor. It was also named Krox-24, TIS8, and ZENK. It was originally discovered in mice.
Function
[edit]The protein encoded by this gene belongs to the EGR family of Cys2His2-type zinc finger proteins. It is a nuclear protein and functions as a transcriptional regulator. The products of target genes it activates are required for differentiation and mitogenesis. Studies suggest this is a tumor suppressor gene.[5]
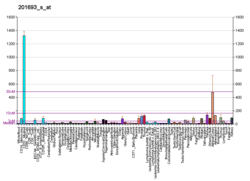
It has a distinct pattern of expression in the brain, and its induction has been shown to be associated with neuronal activity. Several studies suggest it has a role in neuronal plasticity.[6]
EGR-1 is an important transcription factor in memory formation. It has an essential role in brain neuron epigenetic reprogramming. EGR-1 recruits the TET1 protein that initiates a pathway of DNA demethylation.[7] Removing DNA methylation marks allows the activation of downstream genes. EGR-1, together with TET1, is employed in programming the distribution of methylation sites on brain DNA during brain development, in learning and in long-term neuronal plasticity. EGR-1 has also been found to regulate the expression of VAMP2 (a protein important for synaptic exocytosis).[8]
Beside its function in the nervous system, there is significant evidence that EGR-1 along with its paralog EGR-2 is induced in fibrotic diseases has key functions in fibrinogenesis and is necessary for experimentally induced fibrosis in mice.[9]
It may also be involved in ovarian function [10]
Structure
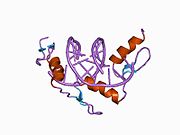
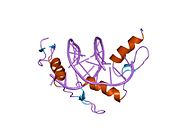
[edit]The DNA-binding domain of EGR-1 consists of three zinc finger domains of the Cys2His2 type. The amino acid structure of the EGR-1 zinc finger domain is given in this table, using the single letter amino acid code. The fingers 1 to 3 are indicated by f1 - f3. The numbers are in reference to the residues (amino acids) of alpha helix (there is no zero). The residues marked 'x' are not part of the zinc fingers, but rather serve to connect them all together.
| -1 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | x | x | x | x | x | |||||||||||||||||||||||
| f1 | M | A | E | E | R | P | Y | A | C | P | V | E | S | C | D | R | R | F | S | R | S | D | E | L | T | R | H | I | R | I | H | T | G | Q | K | P | |
| f2 | F | Q | C | R | I | - | - | C | M | R | N | F | S | R | S | D | H | L | T | T | H | I | R | T | H | T | G | E | K | P | |||||||
| f3 | F | A | C | D | I | - | - | C | G | R | K | F | A | R | S | D | E | R | K | R | H | T | K | I | H | L | R | Q | K | D |
Amino acid key: Alanine (Ala, A), Arginine (Arg, R), Asparagine (Asn, N), Aspartic acid (Asp, D), Cysteine (Cys, C), Glutamic acid (Glu, E), Glutamine (Gln, Q), Glycine (Gly, G), Histidine (His, H), Isoleucine (Ile, I), Leucine (Leu, L), Lysine (Lys, K), Methionine (Met, M), Phenylalanine (Phe, F), Proline (Pro, P), Serine (Ser, S), Threonine (Thr, T), Tryptophan (Trp, W), Tyrosine (Tyr, Y), Valine (Val, V)
The crystal structure of DNA bound by the zinc finger domain of EGR-1 was solved in 1991, which greatly aided early research in zinc finger DNA-binding domains.[11]
The human EGR-1 protein contains (in its unprocessed form) 543 amino acids with a molecular weight of 57.5 kDa, and the gene is located on the chromosome 5.
DNA binding specificity
[edit]EGR-1 binds the DNA sequence 5'-GCG TGG GCG-3' (and similar ones like 5'-GCG GGG GCG-3').[12][13] The f1 position 6 binds the 5' G (the first base count from the left); the f1 position 3 to the second base (C); f1 position -1 binds to the third position (G); f2 position 6 to the fourth base (T); and so on.
Interactions
[edit]EGR-1 has been shown to interact with:
See also
[edit]References
[edit]- ^ a b c GRCh38: Ensembl release 89: ENSG00000120738 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000038418 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Entrez Gene: EGR1 early growth response 1".
- ^ Knapska E, Kaczmarek L (November 2004). "A gene for neuronal plasticity in the mammalian brain: Zif268/Egr-1/NGFI-A/Krox-24/TIS8/ZENK?". Progress in Neurobiology. 74 (4): 183–211. doi:10.1016/j.pneurobio.2004.05.007. PMID 15556287. S2CID 39251786.
- ^ Sun Z, Xu X, He J, Murray A, Sun MA, Wei X, Wang X, McCoig E, Xie E, Jiang X, Li L, Zhu J, Chen J, Morozov A, Pickrell AM, Theus MH, Xie H. EGR1 recruits TET1 to shape the brain methylome during development and upon neuronal activity. Nat Commun. 2019 Aug 29;10(1):3892. doi: 10.1038/s41467-019-11905-3. PMID 31467272
- ^ Petersohn D, Thiel G (August 1996). "Role of zinc-finger proteins Sp1 and zif268/egr-1 in transcriptional regulation of the human synaptobrevin II gene". European Journal of Biochemistry. 239 (3): 827–34. doi:10.1111/j.1432-1033.1996.0827u.x. PMID 8774732.
- ^ Bhattacharyya S, Wu M, Fang F, Tourtellotte W, Feghali-Bostwick C, Varga J (May 2011). "Early growth response transcription factors: key mediators of fibrosis and novel targets for anti-fibrotic therapy". Matrix Biology. 30 (4): 235–42. doi:10.1016/j.matbio.2011.03.005. PMC 3135176. PMID 21511034.
- ^ Han P, Guerrero-Netro H, Estienne A, Cao B, Price CA (October 2017). "Regulation and action of early growth response 1 in bovine granulosa cells". Reproduction. 154 (4): 547–557. doi:10.1530/REP-17-0243. PMID 28733346.
- ^ Pavletich NP, Pabo CO (May 1991). "Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A". Science. 252 (5007): 809–17. doi:10.1126/science.2028256. PMID 2028256. S2CID 38000717.
- ^ Christy B, Nathans D (November 1989). "DNA binding site of the growth factor-inducible protein Zif268". Proceedings of the National Academy of Sciences of the United States of America. 86 (22): 8737–41. Bibcode:1989PNAS...86.8737C. doi:10.1073/pnas.86.22.8737. PMC 298363. PMID 2510170.
- ^ Swirnoff AH, Milbrandt J (April 1995). "DNA-binding specificity of NGFI-A and related zinc finger transcription factors". Molecular and Cellular Biology. 15 (4): 2275–87. doi:10.1128/mcb.15.4.2275. PMC 230455. PMID 7891721.
- ^ Zhang F, Lin M, Abidi P, Thiel G, Liu J (November 2003). "Specific interaction of Egr1 and c/EBPbeta leads to the transcriptional activation of the human low density lipoprotein receptor gene". The Journal of Biological Chemistry. 278 (45): 44246–54. doi:10.1074/jbc.M305564200. PMID 12947119.
- ^ a b Silverman ES, Du J, Williams AJ, Wadgaonkar R, Drazen JM, Collins T (November 1998). "cAMP-response-element-binding-protein-binding protein (CBP) and p300 are transcriptional co-activators of early growth response factor-1 (Egr-1)". The Biochemical Journal. 336 ( Pt 1) (1): 183–9. doi:10.1042/bj3360183. PMC 1219856. PMID 9806899.
- ^ Russo MW, Sevetson BR, Milbrandt J (July 1995). "Identification of NAB1, a repressor of NGFI-A- and Krox20-mediated transcription". Proceedings of the National Academy of Sciences of the United States of America. 92 (15): 6873–7. Bibcode:1995PNAS...92.6873R. doi:10.1073/pnas.92.15.6873. PMC 41432. PMID 7624335.
- ^ Liu J, Grogan L, Nau MM, Allegra CJ, Chu E, Wright JJ (April 2001). "Physical interaction between p53 and primary response gene Egr-1". International Journal of Oncology. 18 (4): 863–70. doi:10.3892/ijo.18.4.863. PMID 11251186.
- ^ Bae MH, Jeong CH, Kim SH, Bae MK, Jeong JW, Ahn MY, et al. (October 2002). "Regulation of Egr-1 by association with the proteasome component C8". Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 1592 (2): 163–7. doi:10.1016/s0167-4889(02)00310-5. PMID 12379479.
Further reading
[edit]- Heath RG (March 1975). "Brain function and behavior. I. Emotion and sensory phenomena in psychotic patients and in experimental animals". The Journal of Nervous and Mental Disease. 160 (3): 159–75. doi:10.1097/00005053-197503000-00002. PMID 1090709. S2CID 23024944.
- Silverman ES, Collins T (March 1999). "Pathways of Egr-1-mediated gene transcription in vascular biology". The American Journal of Pathology. 154 (3): 665–70. doi:10.1016/S0002-9440(10)65312-6. PMC 1866415. PMID 10079243.
- Adamson ED, Mercola D (2002). "Egr1 transcription factor: multiple roles in prostate tumor cell growth and survival". Tumour Biology. 23 (2): 93–102. doi:10.1159/000059711. PMID 12065847. S2CID 46795197.
- Blaschke F, Bruemmer D, Law RE (August 2004). "Egr-1 is a major vascular pathogenic transcription factor in atherosclerosis and restenosis". Reviews in Endocrine & Metabolic Disorders. 5 (3): 249–54. doi:10.1023/B:REMD.0000032413.88756.ee. PMID 15211096. S2CID 11968305.
- Abdulkadir SA (November 2005). "Mechanisms of prostate tumorigenesis: roles for transcription factors Nkx3.1 and Egr1". Annals of the New York Academy of Sciences. 1059 (1): 33–40. Bibcode:2005NYASA1059...33A. doi:10.1196/annals.1339.018. PMID 16382041. S2CID 6774788.
- Khachigian LM (February 2006). "Early growth response-1 in cardiovascular pathobiology". Circulation Research. 98 (2): 186–91. doi:10.1161/01.RES.0000200177.53882.c3. PMID 16456111.
External links
[edit]- Zif+268+protein,+human at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
- FactorBook Egr-1
- Overview of all the structural information available in the PDB for UniProt: P18146 (Human Early growth response protein 1) at the PDBe-KB.
- Overview of all the structural information available in the PDB for UniProt: P08046 (Mouse Early growth response protein 1) at the PDBe-KB.