Valosin-containing protein
Valosin-containing protein (VCP) or transitional endoplasmic reticulum ATPase (TER ATPase) also known as p97 in mammals and CDC48 in S. cerevisiae, is an enzyme that in humans is encoded by the VCP gene.[5][6][7] The TER ATPase is an ATPase enzyme present in all eukaryotes and archaebacteria. Its main function is to segregate protein molecules from large cellular structures such as protein assemblies, organelle membranes and chromatin, and thus facilitate the degradation of released polypeptides by the multi-subunit protease proteasome.
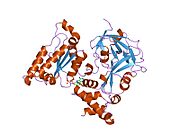
VCP/p97/CDC48 is a member of the AAA+ (extended family of ATPases associated with various cellular activities) ATPase family. Enzymes of this family are found in all species from bacteria to humans. Many of them are important chaperones that regulate folding or unfolding of substrate proteins. VCP is a type II AAA+ ATPase, which means that it contains two tandem ATPase domains (named D1 and D2, respectively) (Figure 1).

The two ATPase domains are connected by a short polypeptide linker. A domain preceding the D1 domain (N-terminal domain) and a short carboxyl-terminal tail are involved in interaction with cofactors.[8] The N-domain is connected to the D1 domain by a short N-D1 linker.
Most known substrates of VCP are modified with ubiquitin chains and degraded by the 26S proteasome. Accordingly, many VCP coenzymes and adaptors have domains that can recognize ubiquitin.[9] It has become evident that the interplays between ubiquitin and VCP cofactors are critical for many of the proposed functions, although the precise role of these interactions remains to be elucidated.
Discovery
[edit]CDC48 was discovered in a genetic screen for genes involved in cell cycle regulation in budding yeast.[10] The screen identified several alleles of Cdc48 that affect cell growth at non-permissive temperatures. A search for the mammalian homolog of CDC48 (valosin) revealed a 97 kDa protein precursor named "valosin-containing protein (VCP)" or p97, and also showed that it was only generated as an artefact of purification rather than during physiological processing.[11] Even without evidence that valosin is a physiological product, the VCP nomenclature continues to be used in the literature.
Tissue and subcelluar distribution
[edit]VCP is one of the most abundant cytoplasmic proteins in eukaryotic cells. It is ubiquitously expressed in all tissues in multicellular organisms. In humans, the mRNA expression of VCP was found to be moderately elevated in certain types of cancer.[9]
In mammalian cells, VCP is predominantly localized to the cytoplasm, and a significant fraction is associated to membranes of cellular organelles such as the endoplasmic reticulum (ER), Golgi, mitochondria, and endosomes.[6][12][13][14][15] The subcellular localization of CDC48 has not been fully characterized, but is likely to be similar to the mammalian counterpart. A fraction of VCP was also found in the nucleus.[16]
Structure
[edit]According to the crystal structures of full-length wild-type VCP,[17][18] six VCP subunits assemble into a barrel-like structure, in which the N-D1 and D2 domains form two concentric, stacked rings (Figure 2).

The N-D1 ring is larger (162 Å in diameter) than the D2 ring (113 Å) due to the laterally attached N-domains. The D1 and D2 domains are highly homologous in both sequence and structure, but they serve distinct functions. For example, the hexameric assembly of VCP only requires the D1 but not the D2 domain.[19] Unlike many bacterial AAA+ proteins, assembly of VCP hexamer does not depend on the presence of nucleotide. The VCP hexameric assembly can undergo dramatic conformational changes during nucleotide hydrolysis cycle,[20][21][22][23][24] and it is generally believed that these conformational changes generate mechanical force, which is applied to substrate molecules to influence their stability and function. However, how precisely VCP generates force is unclear.
The ATP hydrolysis cycle
[edit]The ATP hydrolyzing activity is indispensable for the VCP functions.[25] The two ATPase domains of VCP (D1 and D2) are not equivalent because the D2 domain displays higher ATPase activity than the D1 domain in wild-type protein. Nevertheless, their activities are dependent of each other.[26][27][28][29] For example, nucleotide binding to the D1 domain is required for ATP binding to the D2 domain and nucleotide binding and hydrolysis in D2 is required for the D1 domain to hydrolyze ATP.
The ATPase activity of VCP can be influenced by many factors. For example, it can be stimulated by heat[29] or by a putative substrate protein.[30] In Leishmania infantum, the LiVCP protein is essential for the intracellular development of the parasite and its survival under heat stress.[31] Association with cofactors can have either positive or negative impact on the p97 ATPase activity.[32][33]
Mutations in VCP can also influence its activity. For example, VCP mutant proteins carrying single point mutations found in patients with multisystem proteinopathy (MSP; formerly known as IBMPFD (inclusion body myopathy associated with Paget disease of the bone and frontotemporal dementia)) (see below) have 2-3 fold increase in ATPase activity.[27][34][35]
VCP-interacting proteins
[edit]Recent proteomic studies have identified a large number of p97-interacting proteins. Many of these proteins serve as adaptors that link VCP to a particular subcellular compartment to function in a specific cellular pathway. Others function as adaptors that recruit substrates to VCP for processing. Some VCP-interacting proteins are also enzymes such as N-glycanase, ubiquitin ligase, and deubiquitinase, which assist VCP in processing substrates.
Most cofactors bind VCP through its N-domain, but a few interact with the short carboxy-terminal tail in VCP. Representative proteins interacting with the N-domain are Ufd1, Npl4, p47 and FAF1.[36][37][38] Examples of cofactors that interact with the carboxy-terminal tail of VCP are PLAA, PNGase, and Ufd2.[39][40][41]
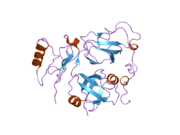
The molecular basis for cofactor binding has been studied for some cofactors that interact with the VCP N-domain. The N-domain consists of two sub-domains of roughly equal size: the N-terminal double Y-barrel and a C-terminal b-barrel (Figure 3).

Structural studies show that many cofactor proteins bind to the N-domain at a cleft formed between the two sub-domains.
Among those that bind to the N-domain of VCP, two most frequently occurring sequence motifs are found: one is called UBX motif (ubiquitin regulatory X)[42] and the other is termed VIM (VCP-interacting motif).[43] The UBX domain is an 80-residue module with a fold highly resembling the structure of ubiquitin. The VCP-interacting motif (VIM) is a linear sequence motif (RX5AAX2R) found in a number of VCP cofactors including gp78,[44] SVIP (small VCP-inhibiting protein)[45] and VIMP (VCP interacting membrane protein).[46] Although the UBX domain uses a surface loop whereas the VIM forms a-helix to bind VCP, both UBX and VIM bind at the same location between the two sub-domains of the N-domain (Figure 3).[47] It was proposed that hierarchical binding to distinct cofactors may be essential for the broad functions of VCP.[48][49]
Function
[edit]VCP performs diverse functions through modulating the stability and thus the activity of its substrates. The general function of VCP is to segregate proteins from large protein assembly or immobile cellular structures such as membranes or chromatin, allowing the released protein molecules to be degraded by the proteasome. The functions of VCP can be grouped into the following three major categories.
Protein quality control
[edit]The best characterized function of VCP is to mediate a network of protein quality control processes in order to maintain protein homeostasis.[50] These include endoplasmic reticulum-associated protein degradation (ERAD) and mitochondria-associated degradation.[14][51] In these processes, ATP hydrolysis by VCP is required to extract aberrant proteins from the membranes of the ER or mitochondria. VCP is also required to release defective translation products stalled on ribosome in a process termed ribosome-associated degradation.[52][53][54] It appears that only after extraction from the membranes or large protein assembly like ribosome, can polypeptides be degraded by the proteasome. In addition to this ‘segregase’ function, VCP might have an additional role in shuttling the released polypeptides to the proteasome. This chaperoning function seems to be particularly important for degradation of certain aggregation-prone misfolded proteins in nucleus.[55] Several lines of evidence also implicate p97 in autophagy, a process that turns over cellular proteins (including misfolded ones) by engulfing them into double-membrane-surrounded vesicles named autophagosome, but the precise role of VCP in this process is unclear.[56]
Chromatin-associated functions
[edit]VCP also functions broadly in eukaryotic nucleus by releasing protein molecules from chromatins in a manner analogous to that in ERAD.[57] The identified VCP substrates include transcriptional repressor α2 and RNA polymerase (Pol) II complex and CMG DNA helicase in budding yeast, and the DNA replicating licensing factor CDT1, DNA repairing proteins DDB2 and XPC, mitosis regulator Aurora B, and certain DNA polymerases in mammalian cells. These substrates link VCP function to gene transcription, DNA replication and repair, and cell cycle progression.
Membrane fusion and trafficking
[edit]Biochemical and genetic studies have also implicated VCP in fusion of vesicles that lead to the formation of Golgi apparatus at the end of mitosis.[58] This process requires the ubiquitin binding adaptor p47 and a p97-associated deubiquitinase VCIP135, and thus connecting membrane fusion to the ubiquitin pathways. However, the precise role of VCP in Golgi formation is unclear due to lack of information on relevant substrate(s). Recent studies also suggest that VCP may regulate vesicle trafficking from plasma membrane to the lysosome, a process termed endocytosis.[56] Antibody fragment-based inhibitors have been developed by a team led by Arkin to inhibit the interaction between p97 and p47, selectively modulating the Golgi reassembly process.[59]
Clinical significance
[edit]Links to human diseases
[edit]Mutations in VCP were first reported to cause a syndrome characterized by frontotemporal dementia, inclusion body myopathy, and Paget's disease of the bone by Virginia Kimonis in 2004.[60] In 2010, mutations in VCP were also found to be a cause of amyotrophic lateral sclerosis by Bryan Traynor and Adriano Chiò.[61] This discovery was notable as it represented an initial genetic link between two disparate neurological diseases, amyotrophic lateral sclerosis and frontotemporal dementia. In 2020, Edward Lee described a distinct hypomorphic mutation in VCP associated with vacuolar tauopathy, a unique subtype of frontotemporal lobar degeneration with tau inclusions.[62]
Mutations in VCP are an example of pleiotropy, where mutations in the same gene give rise to different phenotypes. The term multisystem proteinopathy (MSP) has been coined to describe this particular form of pleiotropy.[63] Although MSP is rare, growing interest in this syndrome derives from the molecular insights the condition provides into the etiological relationship between common age-related degenerative diseases of muscle, bone and brain. It has been estimated that ~50% of MSP may be caused by missense mutations affecting the valosin-containing protein (VCP) gene.[64]
Cancer therapy
[edit]The first p97 inhibitor Eeyarestatin (EerI) was discovered by screening and characterizing compounds that inhibit the degradation of a fluorescence-labeled ERAD substrate.[65][66] The mechanism of VCP inhibition by EerI is unclear, but when applied to cells, it induces biological phenotypes associated with VCP inhibition such as ERAD inhibition, ER stress elevation, and apoptosis induction. Importantly, EerI displays significant cancer-killing activity in vitro preferentially against cancer cells isolated from patients, and it can synergize with the proteasome inhibitor bortezomib to kill cancer cells.[67] These observations prompt the idea of targeting VCP as a potential cancer therapy. This idea was further confirmed by studying several ATP competitive and allosteric inhibitors.[68][69][70] More recently, a potent and specific VCP inhibitor CB-5083 has been developed, which demonstrates promising anti-cancer activities in mouse xenograft tumor models.[71] The compound is now being evaluated in a phase 1 clinical trial.[72]
Notes
[edit]
The 2016 version of this article was updated by an external expert under a dual publication model. The corresponding academic peer reviewed article was published in Gene and can be cited as: Di Xia; Wai Kwan Tang; Yihong Ye (25 May 2016). "Structure and function of the AAA+ ATPase p97/Cdc48p". Gene. Gene Wiki Review Series. 583 (1): 64–77. doi:10.1016/J.GENE.2016.02.042. ISSN 0378-1119. PMC 4821690. PMID 26945625. Wikidata Q36767612. |
References
[edit]- ^ a b c GRCh38: Ensembl release 89: ENSG00000165280 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000028452 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ Druck T, Gu Y, Prabhala G, Cannizzaro LA, Park SH, Huebner K, Keen JH (November 1995). "Chromosome localization of human genes for clathrin adaptor polypeptides AP2 beta and AP50 and the clathrin-binding protein, VCP". Genomics. 30 (1): 94–7. doi:10.1006/geno.1995.0016. PMID 8595912.
- ^ a b Rabouille C, Levine TP, Peters JM, Warren G (September 1995). "An NSF-like ATPase, p97, and NSF mediate cisternal regrowth from mitotic Golgi fragments". Cell. 82 (6): 905–14. doi:10.1016/0092-8674(95)90270-8. PMID 7553851. S2CID 2663141.
- ^ "Entrez Gene: VCP valosin-containing protein".
- ^ Ogura T, Wilkinson AJ (July 2001). "AAA+ superfamily ATPases: common structure--diverse function". Genes to Cells. 6 (7): 575–97. doi:10.1046/j.1365-2443.2001.00447.x. PMID 11473577. S2CID 6740778.
- ^ a b Ye Y (October 2006). "Diverse functions with a common regulator: ubiquitin takes command of an AAA ATPase". Journal of Structural Biology. 156 (1): 29–40. doi:10.1016/j.jsb.2006.01.005. PMID 16529947.
- ^ Moir D, Stewart SE, Osmond BC, Botstein D (April 1982). "Cold-sensitive cell-division-cycle mutants of yeast: isolation, properties, and pseudoreversion studies". Genetics. 100 (4): 547–63. doi:10.1093/genetics/100.4.547. PMC 1201831. PMID 6749598.
- ^ Koller KJ, Brownstein MJ (1987). "Use of a cDNA clone to identify a supposed precursor protein containing valosin". Nature. 325 (6104): 542–5. Bibcode:1987Natur.325..542K. doi:10.1038/325542a0. PMID 3468358. S2CID 19200775.
- ^ Acharya U, Jacobs R, Peters JM, Watson N, Farquhar MG, Malhotra V (September 1995). "The formation of Golgi stacks from vesiculated Golgi membranes requires two distinct fusion events". Cell. 82 (6): 895–904. doi:10.1016/0092-8674(95)90269-4. PMID 7553850. S2CID 14725335.
- ^ Latterich M, Fröhlich KU, Schekman R (September 1995). "Membrane fusion and the cell cycle: Cdc48p participates in the fusion of ER membranes". Cell. 82 (6): 885–93. doi:10.1016/0092-8674(95)90268-6. PMID 7553849. S2CID 17922017.
- ^ a b Xu S, Peng G, Wang Y, Fang S, Karbowski M (February 2011). "The AAA-ATPase p97 is essential for outer mitochondrial membrane protein turnover". Molecular Biology of the Cell. 22 (3): 291–300. doi:10.1091/mbc.E10-09-0748. PMC 3031461. PMID 21118995.
- ^ Ramanathan HN, Ye Y (February 2012). "The p97 ATPase associates with EEA1 to regulate the size of early endosomes". Cell Research. 22 (2): 346–59. doi:10.1038/cr.2011.80. PMC 3271578. PMID 21556036.
- ^ Madeo F, Schlauer J, Zischka H, Mecke D, Fröhlich KU (January 1998). "Tyrosine phosphorylation regulates cell cycle-dependent nuclear localization of Cdc48p". Molecular Biology of the Cell. 9 (1): 131–41. doi:10.1091/mbc.9.1.131. PMC 25228. PMID 9436996.
- ^ DeLaBarre B, Brunger AT (October 2003). "Complete structure of p97/valosin-containing protein reveals communication between nucleotide domains". Nature Structural Biology. 10 (10): 856–63. doi:10.1038/nsb972. PMID 12949490. S2CID 19281416.
- ^ Davies JM, Brunger AT, Weis WI (May 2008). "Improved structures of full-length p97, an AAA ATPase: implications for mechanisms of nucleotide-dependent conformational change". Structure. 16 (5): 715–26. doi:10.1016/j.str.2008.02.010. PMID 18462676.
- ^ Wang Q, Song C, Li CC (January 2003). "Hexamerization of p97-VCP is promoted by ATP binding to the D1 domain and required for ATPase and biological activities". Biochemical and Biophysical Research Communications. 300 (2): 253–60. doi:10.1016/s0006-291x(02)02840-1. PMID 12504076.
- ^ Beuron F, Dreveny I, Yuan X, Pye VE, McKeown C, Briggs LC, Cliff MJ, Kaneko Y, Wallis R, Isaacson RL, Ladbury JE, Matthews SJ, Kondo H, Zhang X, Freemont PS (May 2006). "Conformational changes in the AAA ATPase p97-p47 adaptor complex". The EMBO Journal. 25 (9): 1967–76. doi:10.1038/sj.emboj.7601055. PMC 1456939. PMID 16601695.
- ^ Beuron F, Flynn TC, Ma J, Kondo H, Zhang X, Freemont PS (March 2003). "Motions and negative cooperativity between p97 domains revealed by cryo-electron microscopy and quantised elastic deformational model". Journal of Molecular Biology. 327 (3): 619–29. doi:10.1016/s0022-2836(03)00178-5. PMID 12634057.
- ^ DeLaBarre B, Brunger AT (March 2005). "Nucleotide dependent motion and mechanism of action of p97/VCP". Journal of Molecular Biology. 347 (2): 437–52. doi:10.1016/j.jmb.2005.01.060. PMID 15740751.
- ^ Rouiller I, DeLaBarre B, May AP, Weis WI, Brunger AT, Milligan RA, Wilson-Kubalek EM (December 2002). "Conformational changes of the multifunction p97 AAA ATPase during its ATPase cycle". Nature Structural Biology. 9 (12): 950–7. doi:10.1038/nsb872. PMID 12434150. S2CID 16061425.
- ^ Tang WK, Li D, Li CC, Esser L, Dai R, Guo L, Xia D (July 2010). "A novel ATP-dependent conformation in p97 N-D1 fragment revealed by crystal structures of disease-related mutants". The EMBO Journal. 29 (13): 2217–29. doi:10.1038/emboj.2010.104. PMC 2905243. PMID 20512113.
- ^ Wang Q, Song C, Li CC (2004). "Molecular perspectives on p97-VCP: progress in understanding its structure and diverse biological functions". Journal of Structural Biology. 146 (1–2): 44–57. doi:10.1016/j.jsb.2003.11.014. PMID 15037236.
- ^ Nishikori S, Esaki M, Yamanaka K, Sugimoto S, Ogura T (May 2011). "Positive cooperativity of the p97 AAA ATPase is critical for essential functions". The Journal of Biological Chemistry. 286 (18): 15815–20. doi:10.1074/jbc.M110.201400. PMC 3091191. PMID 21454554.
- ^ a b Tang WK, Xia D (December 2013). "Altered intersubunit communication is the molecular basis for functional defects of pathogenic p97 mutants". The Journal of Biological Chemistry. 288 (51): 36624–35. doi:10.1074/jbc.M113.488924. PMC 3868774. PMID 24196964.
- ^ Ye Y, Meyer HH, Rapoport TA (July 2003). "Function of the p97-Ufd1-Npl4 complex in retrotranslocation from the ER to the cytosol: dual recognition of nonubiquitinated polypeptide segments and polyubiquitin chains". The Journal of Cell Biology. 162 (1): 71–84. doi:10.1083/jcb.200302169. PMC 2172719. PMID 12847084.
- ^ a b Song C, Wang Q, Li CC (February 2003). "ATPase activity of p97-valosin-containing protein (VCP). D2 mediates the major enzyme activity, and D1 contributes to the heat-induced activity". The Journal of Biological Chemistry. 278 (6): 3648–55. doi:10.1074/jbc.M208422200. PMID 12446676.
- ^ DeLaBarre B, Christianson JC, Kopito RR, Brunger AT (May 2006). "Central pore residues mediate the p97/VCP activity required for ERAD". Molecular Cell. 22 (4): 451–62. doi:10.1016/j.molcel.2006.03.036. PMID 16713576.
- ^ Guedes Aguiar B, Padmanabhan PK, Dumas C, Papadopoulou B (June 2018). "Valosin-containing protein VCP/p97 is essential for the intracellular development of Leishmania and its survival under heat stress". Cellular Microbiology. 20 (10): e12867. doi:10.1111/cmi.12867. PMID 29895095. S2CID 48359590.
- ^ Meyer HH, Kondo H, Warren G (October 1998). "The p47 co-factor regulates the ATPase activity of the membrane fusion protein, p97". FEBS Letters. 437 (3): 255–7. doi:10.1016/s0014-5793(98)01232-0. PMID 9824302. S2CID 33962985.
- ^ Zhang X, Gui L, Zhang X, Bulfer SL, Sanghez V, Wong DE, Lee Y, Lehmann L, Lee JS, Shih PY, Lin HJ, Iacovino M, Weihl CC, Arkin MR, Wang Y, Chou TF (April 2015). "Altered cofactor regulation with disease-associated p97/VCP mutations". Proceedings of the National Academy of Sciences of the United States of America. 112 (14): E1705–14. Bibcode:2015PNAS..112E1705Z. doi:10.1073/pnas.1418820112. PMC 4394316. PMID 25775548.
- ^ Halawani D, LeBlanc AC, Rouiller I, Michnick SW, Servant MJ, Latterich M (August 2009). "Hereditary inclusion body myopathy-linked p97/VCP mutations in the NH2 domain and the D1 ring modulate p97/VCP ATPase activity and D2 ring conformation". Molecular and Cellular Biology. 29 (16): 4484–94. doi:10.1128/MCB.00252-09. PMC 2725746. PMID 19506019.
- ^ Weihl CC, Dalal S, Pestronk A, Hanson PI (January 2006). "Inclusion body myopathy-associated mutations in p97/VCP impair endoplasmic reticulum-associated degradation". Human Molecular Genetics. 15 (2): 189–99. doi:10.1093/hmg/ddi426. PMID 16321991.
- ^ Ye Y, Meyer HH, Rapoport TA (December 2001). "The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol". Nature. 414 (6864): 652–6. Bibcode:2001Natur.414..652Y. doi:10.1038/414652a. PMID 11740563. S2CID 23397533.
- ^ Kondo H, Rabouille C, Newman R, Levine TP, Pappin D, Freemont P, Warren G (July 1997). "p47 is a cofactor for p97-mediated membrane fusion". Nature. 388 (6637): 75–8. Bibcode:1997Natur.388R..75K. doi:10.1038/40411. PMID 9214505. S2CID 32646222.
- ^ Song EJ, Yim SH, Kim E, Kim NS, Lee KJ (March 2005). "Human Fas-associated factor 1, interacting with ubiquitinated proteins and valosin-containing protein, is involved in the ubiquitin-proteasome pathway". Molecular and Cellular Biology. 25 (6): 2511–24. doi:10.1128/MCB.25.6.2511-2524.2005. PMC 1061599. PMID 15743842.
- ^ Qiu L, Pashkova N, Walker JR, Winistorfer S, Allali-Hassani A, Akutsu M, Piper R, Dhe-Paganon S (January 2010). "Structure and function of the PLAA/Ufd3-p97/Cdc48 complex". The Journal of Biological Chemistry. 285 (1): 365–72. doi:10.1074/jbc.M109.044685. PMC 2804184. PMID 19887378.
- ^ Zhao G, Zhou X, Wang L, Li G, Schindelin H, Lennarz WJ (May 2007). "Studies on peptide:N-glycanase-p97 interaction suggest that p97 phosphorylation modulates endoplasmic reticulum-associated degradation". Proceedings of the National Academy of Sciences of the United States of America. 104 (21): 8785–90. Bibcode:2007PNAS..104.8785Z. doi:10.1073/pnas.0702966104. PMC 1885580. PMID 17496150.
- ^ Schaeffer V, Akutsu M, Olma MH, Gomes LC, Kawasaki M, Dikic I (May 2014). "Binding of OTULIN to the PUB domain of HOIP controls NF-κB signaling". Molecular Cell. 54 (3): 349–61. doi:10.1016/j.molcel.2014.03.016. PMID 24726327.
- ^ Schuberth C, Buchberger A (August 2008). "UBX domain proteins: major regulators of the AAA ATPase Cdc48/p97". Cellular and Molecular Life Sciences. 65 (15): 2360–71. doi:10.1007/s00018-008-8072-8. PMC 11131665. PMID 18438607.
- ^ Stapf C, Cartwright E, Bycroft M, Hofmann K, Buchberger A (November 2011). "The general definition of the p97/valosin-containing protein (VCP)-interacting motif (VIM) delineates a new family of p97 cofactors". The Journal of Biological Chemistry. 286 (44): 38670–8. doi:10.1074/jbc.M111.274472. PMC 3207395. PMID 21896481.
- ^ Ballar P, Shen Y, Yang H, Fang S (November 2006). "The role of a novel p97/valosin-containing protein-interacting motif of gp78 in endoplasmic reticulum-associated degradation". The Journal of Biological Chemistry. 281 (46): 35359–68. doi:10.1074/jbc.M603355200. PMID 16987818.
- ^ Ballar P, Zhong Y, Nagahama M, Tagaya M, Shen Y, Fang S (November 2007). "Identification of SVIP as an endogenous inhibitor of endoplasmic reticulum-associated degradation". The Journal of Biological Chemistry. 282 (47): 33908–14. doi:10.1074/jbc.M704446200. PMID 17872946.
- ^ Ye Y, Shibata Y, Yun C, Ron D, Rapoport TA (June 2004). "A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol". Nature. 429 (6994): 841–7. Bibcode:2004Natur.429..841Y. doi:10.1038/nature02656. PMID 15215856. S2CID 4317750.
- ^ Hänzelmann P, Schindelin H (November 2011). "The structural and functional basis of the p97/valosin-containing protein (VCP)-interacting motif (VIM): mutually exclusive binding of cofactors to the N-terminal domain of p97". The Journal of Biological Chemistry. 286 (44): 38679–90. doi:10.1074/jbc.M111.274506. PMC 3207442. PMID 21914798.
- ^ Meyer HH, Shorter JG, Seemann J, Pappin D, Warren G (May 2000). "A complex of mammalian ufd1 and npl4 links the AAA-ATPase, p97, to ubiquitin and nuclear transport pathways". The EMBO Journal. 19 (10): 2181–92. doi:10.1093/emboj/19.10.2181. PMC 384367. PMID 10811609.
- ^ Buchberger A, Schindelin H, Hänzelmann P (September 2015). "Control of p97 function by cofactor binding". FEBS Letters. 589 (19 Pt A): 2578–89. doi:10.1016/j.febslet.2015.08.028. PMID 26320413. S2CID 41082524.
- ^ Meyer H, Bug M, Bremer S (February 2012). "Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system". Nature Cell Biology. 14 (2): 117–23. doi:10.1038/ncb2407. PMID 22298039. S2CID 23562362.
- ^ Christianson JC, Ye Y (April 2014). "Cleaning up in the endoplasmic reticulum: ubiquitin in charge". Nature Structural & Molecular Biology. 21 (4): 325–35. doi:10.1038/nsmb.2793. PMC 9397582. PMID 24699081. S2CID 43665193.
- ^ Brandman O, Stewart-Ornstein J, Wong D, Larson A, Williams CC, Li GW, Zhou S, King D, Shen PS, Weibezahn J, Dunn JG, Rouskin S, Inada T, Frost A, Weissman JS (November 2012). "A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress". Cell. 151 (5): 1042–54. doi:10.1016/j.cell.2012.10.044. PMC 3534965. PMID 23178123.
- ^ Defenouillère Q, Yao Y, Mouaikel J, Namane A, Galopier A, Decourty L, Doyen A, Malabat C, Saveanu C, Jacquier A, Fromont-Racine M (March 2013). "Cdc48-associated complex bound to 60S particles is required for the clearance of aberrant translation products". Proceedings of the National Academy of Sciences of the United States of America. 110 (13): 5046–51. Bibcode:2013PNAS..110.5046D. doi:10.1073/pnas.1221724110. PMC 3612664. PMID 23479637.
- ^ Verma R, Oania RS, Kolawa NJ, Deshaies RJ (January 2013). "Cdc48/p97 promotes degradation of aberrant nascent polypeptides bound to the ribosome". eLife. 2: e00308. doi:10.7554/eLife.00308. PMC 3552423. PMID 23358411.
- ^ Gallagher PS, Clowes Candadai SV, Gardner RG (May 2014). "The requirement for Cdc48/p97 in nuclear protein quality control degradation depends on the substrate and correlates with substrate insolubility". Journal of Cell Science. 127 (Pt 9): 1980–91. doi:10.1242/jcs.141838. PMC 4004975. PMID 24569878.
- ^ a b Bug M, Meyer H (August 2012). "Expanding into new markets--VCP/p97 in endocytosis and autophagy". Journal of Structural Biology. 179 (2): 78–82. doi:10.1016/j.jsb.2012.03.003. PMID 22450227.
- ^ Dantuma NP, Acs K, Luijsterburg MS (November 2014). "Should I stay or should I go: VCP/p97-mediated chromatin extraction in the DNA damage response". Experimental Cell Research. 329 (1): 9–17. doi:10.1016/j.yexcr.2014.08.025. PMID 25169698.
- ^ Uchiyama K, Kondo H (February 2005). "p97/p47-Mediated biogenesis of Golgi and ER". Journal of Biochemistry. 137 (2): 115–9. doi:10.1093/jb/mvi028. PMID 15749824. S2CID 10459261.
- ^ Jiang, Ziwen; Kuo, Yu-Hsuan; Zhong, Mengqi; Zhang, Jianchao; Zhou, Xin X.; Xing, Lijuan; Wells, James A.; Wang, Yanzhuang; Arkin, Michelle R. (2022-07-27). "Adaptor-Specific Antibody Fragment Inhibitors for the Intracellular Modulation of p97 (VCP) Protein–Protein Interactions". Journal of the American Chemical Society. 144 (29): 13218–13225. doi:10.1021/jacs.2c03665. ISSN 0002-7863. PMC 9335864. PMID 35819848.
- ^ Watts, Giles D. J.; Wymer, Jill; Kovach, Margaret J.; Mehta, Sarju G.; Mumm, Steven; Darvish, Daniel; Pestronk, Alan; Whyte, Michael P.; Kimonis, Virginia E. (2004). "Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein". Nature Genetics. 36 (4): 377–381. doi:10.1038/ng1332. ISSN 1061-4036. PMID 15034582.
- ^ Johnson, Janel O.; Mandrioli, Jessica; Benatar, Michael; Abramzon, Yevgeniya; Van Deerlin, Vivianna M.; Trojanowski, John Q.; Gibbs, J. Raphael; Brunetti, Maura; Gronka, Susan (2010-12-09). "Exome sequencing reveals VCP mutations as a cause of familial ALS". Neuron. 68 (5): 857–864. doi:10.1016/j.neuron.2010.11.036. ISSN 1097-4199. PMC 3032425. PMID 21145000.
- ^ Darwich, N.F., Phan J.M.; et al. (2020). "Autosomal dominant VCP hypomorph mutation impairs disaggregation of PHF-tau". Science. 370 (6519): eaay8826. doi:10.1126/science.aay8826. PMC 7818661. PMID 33004675.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Taylor, J. Paul (2015-08-25). "Multisystem proteinopathy: intersecting genetics in muscle, bone, and brain degeneration". Neurology. 85 (8): 658–660. doi:10.1212/WNL.0000000000001862. ISSN 1526-632X. PMID 26208960. S2CID 42203997.
- ^ Le Ber I, Van Bortel I, Nicolas G, Bouya-Ahmed K, Camuzat A, Wallon D, De Septenville A, Latouche M, Lattante S, Kabashi E, Jornea L, Hannequin D, Brice A (April 2014). "hnRNPA2B1 and hnRNPA1 mutations are rare in patients with "multisystem proteinopathy" and frontotemporal lobar degeneration phenotypes". Neurobiology of Aging. 35 (4): 934.e5–6. doi:10.1016/j.neurobiolaging.2013.09.016. PMID 24119545. S2CID 207160856.
- ^ Fiebiger E, Hirsch C, Vyas JM, Gordon E, Ploegh HL, Tortorella D (April 2004). "Dissection of the dislocation pathway for type I membrane proteins with a new small molecule inhibitor, eeyarestatin". Molecular Biology of the Cell. 15 (4): 1635–46. doi:10.1091/mbc.E03-07-0506. PMC 379262. PMID 14767067.
- ^ Wang Q, Shinkre BA, Lee JG, Weniger MA, Liu Y, Chen W, Wiestner A, Trenkle WC, Ye Y (November 2010). "The ERAD inhibitor Eeyarestatin I is a bifunctional compound with a membrane-binding domain and a p97/VCP inhibitory group". PLOS ONE. 5 (11): e15479. Bibcode:2010PLoSO...515479W. doi:10.1371/journal.pone.0015479. PMC 2993181. PMID 21124757.
- ^ Wang Q, Mora-Jensen H, Weniger MA, Perez-Galan P, Wolford C, Hai T, Ron D, Chen W, Trenkle W, Wiestner A, Ye Y (February 2009). "ERAD inhibitors integrate ER stress with an epigenetic mechanism to activate BH3-only protein NOXA in cancer cells". Proceedings of the National Academy of Sciences of the United States of America. 106 (7): 2200–5. Bibcode:2009PNAS..106.2200W. doi:10.1073/pnas.0807611106. PMC 2629785. PMID 19164757.
- ^ Chou TF, Li K, Frankowski KJ, Schoenen FJ, Deshaies RJ (February 2013). "Structure-activity relationship study reveals ML240 and ML241 as potent and selective inhibitors of p97 ATPase". ChemMedChem. 8 (2): 297–312. doi:10.1002/cmdc.201200520. PMC 3662613. PMID 23316025.
- ^ Chou TF, Brown SJ, Minond D, Nordin BE, Li K, Jones AC, Chase P, Porubsky PR, Stoltz BM, Schoenen FJ, Patricelli MP, Hodder P, Rosen H, Deshaies RJ (March 2011). "Reversible inhibitor of p97, DBeQ, impairs both ubiquitin-dependent and autophagic protein clearance pathways". Proceedings of the National Academy of Sciences of the United States of America. 108 (12): 4834–9. Bibcode:2011PNAS..108.4834C. doi:10.1073/pnas.1015312108. PMC 3064330. PMID 21383145.
- ^ Magnaghi P, D'Alessio R, Valsasina B, Avanzi N, Rizzi S, Asa D, Gasparri F, Cozzi L, Cucchi U, Orrenius C, Polucci P, Ballinari D, Perrera C, Leone A, Cervi G, Casale E, Xiao Y, Wong C, Anderson DJ, Galvani A, Donati D, O'Brien T, Jackson PK, Isacchi A (September 2013). "Covalent and allosteric inhibitors of the ATPase VCP/p97 induce cancer cell death". Nature Chemical Biology. 9 (9): 548–56. doi:10.1038/nchembio.1313. PMID 23892893.
- ^ Anderson DJ, Le Moigne R, Djakovic S, Kumar B, Rice J, Wong S, Wang J, Yao B, Valle E, Kiss von Soly S, Madriaga A, Soriano F, Menon MK, Wu ZY, Kampmann M, Chen Y, Weissman JS, Aftab BT, Yakes FM, Shawver L, Zhou HJ, Wustrow D, Rolfe M (November 2015). "Targeting the AAA ATPase p97 as an Approach to Treat Cancer through Disruption of Protein Homeostasis". Cancer Cell. 28 (5): 653–665. doi:10.1016/j.ccell.2015.10.002. PMC 4941640. PMID 26555175.
- ^ Zhou HJ, Wang J, Yao B, Wong S, Djakovic S, Kumar B, Rice J, Valle E, Soriano F, Menon MK, Madriaga A, Kiss von Soly S, Kumar A, Parlati F, Yakes FM, Shawver L, Le Moigne R, Anderson DJ, Rolfe M, Wustrow D (December 2015). "Discovery of a First-in-Class, Potent, Selective, and Orally Bioavailable Inhibitor of the VCP AAA ATPase (CB-5083)". Journal of Medicinal Chemistry. 58 (24): 9480–97. doi:10.1021/acs.jmedchem.5b01346. PMID 26565666.
Further reading
[edit]- Guinto JB, Ritson GP, Taylor JP, Forman MS (July 2007). "Valosin-containing protein and the pathogenesis of frontotemporal dementia associated with inclusion body myopathy". Acta Neuropathologica. 114 (1): 55–61. doi:10.1007/s00401-007-0224-7. PMID 17457594. S2CID 2094590.
- Dawson SJ, White LA (May 1992). "Treatment of Haemophilus aphrophilus endocarditis with ciprofloxacin". The Journal of Infection. 24 (3): 317–20. doi:10.1016/S0163-4453(05)80037-4. PMID 1602151.
- Pleasure IT, Black MM, Keen JH (September 1993). "Valosin-containing protein, VCP, is a ubiquitous clathrin-binding protein". Nature. 365 (6445): 459–62. Bibcode:1993Natur.365..459P. doi:10.1038/365459a0. PMID 8413590. S2CID 4307576.
- Germain-Lee EL, Obie C, Valle D (August 1997). "NVL: a new member of the AAA family of ATPases localized to the nucleus". Genomics. 44 (1): 22–34. doi:10.1006/geno.1997.4856. PMID 9286697.
- Hoyle J, Tan KH, Fisher EM (October 1997). "Mapping the valosin-containing protein (VCP) gene on human chromosome 9 and mouse chromosome 4, and a likely pseudogene on the mouse X chromosome". Mammalian Genome. 8 (10): 778–80. doi:10.1007/s003359900566. PMID 9321476. S2CID 563437.
- Dai RM, Chen E, Longo DL, Gorbea CM, Li CC (February 1998). "Involvement of valosin-containing protein, an ATPase Co-purified with IkappaBalpha and 26 S proteasome, in ubiquitin-proteasome-mediated degradation of IkappaBalpha". The Journal of Biological Chemistry. 273 (6): 3562–73. doi:10.1074/jbc.273.6.3562. PMID 9452483.
- Rabouille C, Kondo H, Newman R, Hui N, Freemont P, Warren G (March 1998). "Syntaxin 5 is a common component of the NSF- and p97-mediated reassembly pathways of Golgi cisternae from mitotic Golgi fragments in vitro". Cell. 92 (5): 603–10. doi:10.1016/S0092-8674(00)81128-9. PMID 9506515. S2CID 17285800.
- Zhang SH, Liu J, Kobayashi R, Tonks NK (June 1999). "Identification of the cell cycle regulator VCP (p97/CDC48) as a substrate of the band 4.1-related protein-tyrosine phosphatase PTPH1". The Journal of Biological Chemistry. 274 (25): 17806–12. doi:10.1074/jbc.274.25.17806. PMID 10364224.
- Zhang H, Wang Q, Kajino K, Greene MI (May 2000). "VCP, a weak ATPase involved in multiple cellular events, interacts physically with BRCA1 in the nucleus of living cells". DNA and Cell Biology. 19 (5): 253–63. doi:10.1089/10445490050021168. PMID 10855792.
- Lavoie C, Chevet E, Roy L, Tonks NK, Fazel A, Posner BI, Paiement J, Bergeron JJ (December 2000). "Tyrosine phosphorylation of p97 regulates transitional endoplasmic reticulum assembly in vitro". Proceedings of the National Academy of Sciences of the United States of America. 97 (25): 13637–42. Bibcode:2000PNAS...9713637L. doi:10.1073/pnas.240278097. PMC 17628. PMID 11087817.
- Seigneurin-Berny D, Verdel A, Curtet S, Lemercier C, Garin J, Rousseaux S, Khochbin S (December 2001). "Identification of components of the murine histone deacetylase 6 complex: link between acetylation and ubiquitination signaling pathways". Molecular and Cellular Biology. 21 (23): 8035–44. doi:10.1128/MCB.21.23.8035-8044.2001. PMC 99970. PMID 11689694.
- Yang CS, Weiner H (April 2002). "Yeast two-hybrid screening identifies binding partners of human Tom34 that have ATPase activity and form a complex with Tom34 in the cytosol". Archives of Biochemistry and Biophysics. 400 (1): 105–10. doi:10.1006/abbi.2002.2778. PMID 11913976.
- Asai T, Tomita Y, Nakatsuka S, Hoshida Y, Myoui A, Yoshikawa H, Aozasa K (March 2002). "VCP (p97) regulates NFkappaB signaling pathway, which is important for metastasis of osteosarcoma cell line". Japanese Journal of Cancer Research. 93 (3): 296–304. doi:10.1111/j.1349-7006.2002.tb02172.x. PMC 5926968. PMID 11927012.
- Kobayashi T, Tanaka K, Inoue K, Kakizuka A (December 2002). "Functional ATPase activity of p97/valosin-containing protein (VCP) is required for the quality control of endoplasmic reticulum in neuronally differentiated mammalian PC12 cells". The Journal of Biological Chemistry. 277 (49): 47358–65. doi:10.1074/jbc.M207783200. PMID 12351637.
- Uchiyama K, Jokitalo E, Kano F, Murata M, Zhang X, Canas B, Newman R, Rabouille C, Pappin D, Freemont P, Kondo H (December 2002). "VCIP135, a novel essential factor for p97/p47-mediated membrane fusion, is required for Golgi and ER assembly in vivo". The Journal of Cell Biology. 159 (5): 855–66. doi:10.1083/jcb.200208112. PMC 2173386. PMID 12473691.
External links
[edit]- GeneReviews/NIH/NCBI/UW entry on Inclusion Body Myopathy with Paget Disease of Bone and/or Frontotemporal Dementia
- Overview of all the structural information available in the PDB for UniProt: P55072 (Transitional endoplasmic reticulum ATPase) at the PDBe-KB.