Ribonuclease T1
| Ribonuclease T1 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 | |||||||||
| Identifiers | |||||||||
| EC no. | 4.6.1.24 | ||||||||
| CAS no. | 9026-12-4 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
| Ribonuclease T1 | |||||||
|---|---|---|---|---|---|---|---|
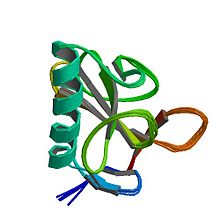 Ribonuclease T1 from Aspergillus oryzae.[1] | |||||||
| Identifiers | |||||||
| Symbol | rntA | ||||||
| PDB | 1YGW | ||||||
| UniProt | P00651 | ||||||
| Other data | |||||||
| EC number | 4.6.1.24 | ||||||
| |||||||
Ribonuclease T1 (EC 4.6.1.24, guanyloribonuclease, Aspergillus oryzae ribonuclease, RNase N1, RNase N2, ribonuclease N3, ribonuclease U1, ribonuclease F1, ribonuclease Ch, ribonuclease PP1, ribonuclease SA, RNase F1, ribonuclease C2, binase, RNase Sa, guanyl-specific RNase, RNase G, RNase T1, ribonuclease guaninenucleotido-2'-transferase (cyclizing), ribonuclease N3, ribonuclease N1) is a fungal endonuclease that cleaves single-stranded RNA after guanine residues, i.e., on their 3' end; the most commonly studied form of this enzyme is the version found in the mold Aspergillus oryzae. Owing to its specificity for guanine, RNase T1 is often used to digest denatured RNA prior to sequencing. Similar to other ribonucleases such as barnase and RNase A, ribonuclease T1 has been popular for folding studies.[2]
Structurally, ribonuclease T1 is a small α+β protein (104 amino acids) with a four-stranded, antiparallel beta sheet covering a long alpha helix (almost five turns). RNase T1 has two disulfide bonds, Cys2-Cys10 and Cys6-Cys103, of which the latter contributes more to its folding stability;[3] complete reduction of both disulfides usually unfolds the protein, although its folding can be rescued with high salt concentrations.[4]
RNase T1 also has four prolines, two of which (Pro39 and Pro55) have cis isomers of their X-Pro peptide bonds. Nonnative isomers of these prolines can retard conformational folding dramatically,[5] folding on a characteristic time scale of 7,000 seconds (almost two hours) at 10 °C and pH 5.[6]
References
[edit]- ^ PDB: 1ygw; Pfeiffer S, Karimi-Nejad Y, Rüterjans H (1997). "Limits of NMR structure determination using variable target function calculations: ribonuclease T1, a case study". Journal of Molecular Biology. 266 (2): 400–423. doi:10.1006/jmbi.1996.0784. PMID 9047372.
- ^ Pace CN, Heinemann U, Hahn U, Saenger W (1991). "Ribonuclease T1: Structure, Function, and Stability". Angewandte Chemie. 30 (4): 343–360. doi:10.1002/anie.199103433.
- ^ Pace CN, Grimsley GR, Thomson JA, Barnett BJ (1988). "Conformational stability and activity of ribonuclease T1 with zero, one, and two intact disulfide bonds". Journal of Biological Chemistry. 263 (24): 11820–11825. doi:10.1016/S0021-9258(18)37859-1. PMID 2457027.
- ^ Oobatake M, Takahashi S, Ooi T (1979). "Conformational stability of ribonuclease T1. II. Salt-induced renaturation". Journal of Biochemistry. 86: 65–70.
- ^ Mayr LM, Odefey CO, Schutkowski M, Schmid FX (1996). "Kinetic analysis of the unfolding and refolding of ribonuclease T1 by a stopped-flow double-mixing technique". Biochemistry. 35 (17): 5550–5561. doi:10.1021/bi953035y. PMID 8611546.
- ^ Mullins LS, Pace CN, Raushel FM (1997). "Conformational stability of ribonuclease T1 measured by hydrogen-deuterium exchange". Protein Science. 6 (7): 1387–1395. doi:10.1002/pro.5560060702. PMC 2143755. PMID 9232639.
External links
[edit]- Ribonuclease+T1 at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
