Topoisomerase
| DNA Topoisomerase, ATP-dependent (type II) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC no. | 5.6.2.2 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| |||||||||
DNA topoisomerases (or topoisomerases) are enzymes that catalyze changes in the topological state of DNA, interconverting relaxed and supercoiled forms, linked (catenated) and unlinked species, and knotted and unknotted DNA.[1][2] Topological issues in DNA arise due to the intertwined nature of its double-helical structure, which, for example, can lead to overwinding of the DNA duplex during DNA replication and transcription. If left unchanged, this torsion would eventually stop the DNA or RNA polymerases involved in these processes from continuing along the DNA helix. A second topological challenge results from the linking or tangling of DNA during replication. Left unresolved, links between replicated DNA will impede cell division. The DNA topoisomerases prevent and correct these types of topological problems. They do this by binding to DNA and cutting the sugar-phosphate backbone of either one (type I topoisomerases) or both (type II topoisomerases) of the DNA strands. This transient break allows the DNA to be untangled or unwound, and, at the end of these processes, the DNA backbone is resealed. Since the overall chemical composition and connectivity of the DNA do not change, the DNA substrate and product are chemical isomers, differing only in their topology.
Discovery
[edit]The first DNA topoisomerase was discovered in bacteria by James C. Wang in 1971 and was initially named ω (omega) protein;[3] it is now called Escherichia coli (E. coli) topoisomerase I (topo I) and is a representative of the type IA family of enzymes. Subsequently, a similar activity was found in eukaryotic cells (rat liver) by James Champoux and Renato Dulbecco;[4] the enzyme responsible, eukaryotic topo I, has a distinct mechanism and is representative of the type IB family. The first type II topoisomerase to be discovered was DNA gyrase from bacteria, by Martin Gellert and coworkers in 1976,[5] and also characterized by Nicholas Cozzarelli and co-workers.[6] DNA gyrase catalyzes the introduction of negative supercoils into DNA and is the only type II enzyme to do this, all the others catalyze DNA relaxation. Type II enzymes are mechanistically distinct from type I in being ATP-dependent and transiently cleaving both DNA strands rather than just one. Type II topoisomerases were subsequently identified from bacterial viruses and eukaryotes.[7][8][9] Topo EC-codes are as follows: ATP-independent (type I), EC 5.6.2.1; ATP-dependent (type II): EC 5.6.2.2. The exception among the type I topoisomerases, reverse gyrase, which contains a helicase domain (EC 3.6.4.12) and introduces positive supercoiling in an ATP-dependent manner. Therefore it is the sole type I topoisomerase classified as EC 5.6.2.2 (Table 1).
DNA topology
[edit]The double-helical structure of DNA involves the intertwining of the two polynucleotide strands around each other, which potentially gives rise to topological problems. DNA topology refers to the crossing of the two DNA strands that alters the twist of the double helix and gives rise to tertiary conformations of DNA, such as supercoils, knots and catenanes.[10] Potential topological issues associated with the double-helical structure of DNA were recognized soon after its structure was first elucidated in 1953 by James Watson, Francis Crick and Rosalind Franklin[11][12][13] and developed further by the work of Max Delbruck and John Cairns.[14][15] Closed-circular double-stranded DNA can be described by 3 parameters: Linking number (Lk), Twist (Tw) and Writhe (Wr) (Fig. 1). Where Lk refers to the number of times the two strands are linked, Tw refers to the number of helical turns in the DNA, measured relative to the helical axis, and Wr quantifies the coiling of the path of the DNA helix in space and is often equated with 'supercoiling'.

The 3 parameters are related as follows: Lk = Tw +Wr. This is a mathematical identity originally obtained by Călugăreanu in 1959[16] and is referred to as the Călugăreanu, or Călugăreanu–White–Fuller, theorem.[17][18] Lk cannot be altered without breaking one or both strands of the helix; Tw and Wr are interconvertible and depend upon the solution conditions. Supercoiling is a vernacular term for DNA with a non-zero linking difference, more correctly referred to as specific linking difference (σ = ΔLk/Lk0, where Lk0 is the mean linking number of the relaxed DNA circle). DNA is said to be positively supercoiled if Lk of it is higher than Lk0 for the relaxed state (Lk-Lko = ΔLk, ΔLk>0); that means that Tw and/or Wr are increased relative to the relaxed molecule. Conversely, DNA is negatively supercoiled if Lk of the molecule is lower than the Lk0 (ΔLk<0).
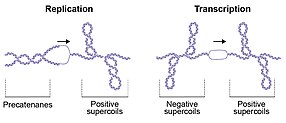
The consequences of topological perturbations in DNA are exemplified by DNA replication during which the strands of the duplex are separated; this separation leads to the formation of positive supercoils (DNA overwinding or overtwisting) ahead of the replication fork and intertwining of the daughter strands (precatenanes) behind[10][19] (Fig. 2). If the positive supercoils are not relaxed, progression of the replication fork is impeded, whereas failure to unlink the daughter strands prevents genome segregation, which is required for cell division.[20] Transcription by RNA polymerase also generates positive supercoiling ahead of, and negative supercoiling behind, the transcriptional complex (Fig. 2). This effect is known as the twin-supercoiled domain model, as described by Leroy Liu and James Wang in 1987.[21] These topological perturbations must be resolved for DNA metabolism to proceed, allowing the cell to efficiently replicate, transcribe and partition the genome to enable cellular division and vitality. Knots in DNA can be found in bacteriophages and as products of recombination reactions.[10] In general, knots in DNA are detrimental and need to be removed (by topoisomerases). DNA catenanes are formed upon the replication of circular molecules and need to be resolved by topoisomerases or recombinases to allow proper separation of daughter molecules during cell division. In addition to the detrimental aspects of DNA topology that require resolution, there are also beneficial aspects. For example, plasmid replication requires negative supercoiling of the origin, which facilitates local melting and exposes single-stranded DNA required to initiate replication. Similarly, initiation of replication from the main bacterial origin oriC also requires negative supercoiling.[22][23] Furthermore, compaction of the E. coli genome is achieved in part by negative supercoiling.
Types
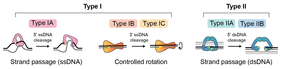
[edit]DNA topoisomerases are enzymes that have evolved to resolve topological problems in DNA (Table 2).[10] They do this via transient breakage of one or both strands of DNA. This has led to the classification of topos into two types: type I, which catalyze reactions involving transient single-stranded breaks, and type II, which catalyze reactions involving transient double-stranded breaks (Fig. 3; Table 2). Sub-types exist within these classifications.
Type I
[edit]These enzymes catalyze changes in DNA topology via transient single-stranded breaks in DNA. Reactions can occur on both single- and double-stranded DNA substrates and can proceed via a 'swivel' or 'strand-passage' mechanism (Fig. 3). The range of reactions includes: DNA supercoil relaxation, unknotting of single-stranded circles, and decatenation, provided at least one partner has a single-stranded region. In the case of the archaeal enzyme, reverse gyrase, positive supercoiling of DNA is possible.[24]
Type IA
[edit]Type IA are monomeric and bind to single-stranded segments of DNA. They introduce a transient single-stranded break through the formation of a tyrosyl-phosphate bond between a tyrosine in the enzyme and a 5′-phosphate in the DNA. The segment of DNA within which the break occurs is called the 'gate' or G-segment, and its cleavage allows the passage of another segment of DNA, the 'transport' or T-segment, to be passed through in a 'strand-passage' process.[25] This is followed by ligation of the G-segment. For strand passage to occur, topo IA must undergo a conformational change to open the DNA gate and allow T-segment transfer. During a DNA relaxation reaction this process changes the linking number of the DNA by +/-1 (Fig. 4). Examples of type IA topoisomerases include prokaryotic topo I and III, eukaryotic topo IIIα and IIIβ and the archaeal enzyme reverse gyrase. Reverse gyrase, which occurs in thermophilic archaea, comprises a type IA topo coupled to a helicase, and is the only known enzyme that can introduce positive supercoils into DNA.[24] The gene encoding reverse gyrase is also found in some groups of thermophilic bacteria, where it was likely transferred by horizontal gene transfer from Archaea.[26]

Type IB
[edit]Type IB topoisomerases catalyze reactions involving transient single-stranded breaks in DNA through the formation of a tyrosyl-phosphate bond between a tyrosine in the enzyme and a 3′-phosphate in the DNA. Rather than utilizing a strand-passage mechanism, these enzymes operate via a 'swivel' or 'controlled rotation' of the cleaved strand around the intact strand.[27] This controlled-rotation mechanism was first described for Vaccinia topo I[27][28] and permits DNA rotation of the free end around the intact strand, the speed being controlled by 'friction' within the enzyme cavity, before the nick is re-ligated (Fig. 3). This results in a variable change of linking number per cleavage and religation event. This mechanism is distinct from that of the type IA enzymes, and the two groups of enzymes are structurally and evolutionarily unrelated. Examples of type IB topoisomerases include eukaryotic nuclear and mitochondrial topo I in addition to viral topo I, though they have been identified in all three domains of life.
Type IC
[edit]Type IC topoisomerases share a similar mechanism to the type IB enzymes but are structurally distinct. The sole representative is topo V, found in the hyperthermophile Methanopyrus kandleri.[29]
Type II
[edit]Type II topoisomerases catalyze changes in DNA topology via transient double-stranded breaks in DNA. Reactions occur on double-stranded DNA substrates and proceed via a strand-passage mechanism (Fig. 5). The range of reactions include DNA relaxation, DNA supercoiling, unknotting, and decatenation. Whereas all type II topoisomerases can catalyze DNA relaxation, gyrase, an archetypal bacterial topoisomerase, can also introduce negative supercoils. In contrast to type I topoisomerases that are generally monomeric, type II topoisomerases are homodimers or heterotetramers. They are classified into two subtypes based on evolutionary, structural, and mechanistic considerations. The general strand-passage mechanism for the type II topos begins with the binding of one DNA duplex, termed the gate segment (G-segment), at the DNA gate. Another duplex, termed the transport segment (T-segment), is captured by an ATP-operated clamp and passed through a transient break in the G-segment, involving 5ʹ phosphotyrosine linkages in both strands, before it is released through the C-gate and the G-segment is re-ligated (Fig. 5). Enzyme turnover requires the binding and hydrolysis of ATP.

Type IIA
[edit]Type IIA topoisomerases catalyze transient double-stranded breaks in DNA through the formation of tyrosyl-phosphate bonds between tyrosines in the enzyme (one on each subunit) and 5′-phosphates staggered by 4 bases in opposite DNA strands. The strand-passage reaction can be intra- or intermolecular (Fig. 5), thus permitting changes in supercoiling and knotting, or unlinking, respectively. This process changes the linking number of the DNA by +/-2. Examples of type IIA topoisomerases include eukaryotic topo IIα and topo IIβ, in addition to bacterial gyrase and topo IV. DNA gyrase conforms to the same double-strand passage mechanism as other type II enzymes but has unique features connected with its ability to introduce negative supercoils into DNA. The G segment is part of a much longer piece of DNA (>100 bp) that is wrapped around the enzyme, one arm of which forms the T-segment that is passed through the double-stranded break (Fig. 5). In the case of gyrase, a substantial amount of the free energy from ATP hydrolysis is transduced into torsional stress in DNA, i.e. supercoiling is an energy-requiring process.[30] Further, in the absence of ATP, gyrase is able to remove negative supercoils in a slower DNA relaxation reaction.
Type IIB
[edit]Type IIB also catalyze transient double-stranded breaks through the formation of tyrosyl-phosphate bonds between tyrosines in the enzyme and 5′-phosphates in opposite strands of the DNA, but in the case of IIB enzymes the double-stranded breaks have a 2-base stagger. Type IIB enzymes show important structural differences, but are evolutionarily related to the type IIA enzymes. These differences include the lack of one of the protein 'gates' (the C gate) (Fig. 5). Originally found in archaea, they have also been found in eukaryotes, and, in particular, in plants; examples include topo VI and topo VIII. Topo VI is the best-studied enzyme of this sub-type and is thought to be a preferential decatenase.[31]
As drug targets
[edit]For the non-specialist perhaps the most important aspect of topoisomerases is their role as drug targets both for antibacterial and anti-cancer chemotherapy; several topoisomerase-targeted antibacterial and anti-cancer drugs are listed among the 2019 World Health Organization Model List of essential Medicines. The reason for this prominence is that their reactions proceed via transient breaks in DNA, which, if stabilized by drug binding, can lead to cell death due to the generation of toxic single- or double-stranded breaks in genomic DNA. The majority of topo-targeted drugs act in this way, i.e. they stabilize the enzyme-DNA covalent cleavage intermediate.[32][33][34]
Antibacterial compounds
[edit]Although type I topos, such as bacterial topo I, are viable antibiotic targets,[35] there are currently no compounds in clinical use that target these enzymes. However, the type II enzymes, DNA gyrase and DNA topoisomerase IV, have enjoyed enormous success as targets for the widely-used fluoroquinolone antibiotics, (Fig. 6).
Fluoroquinolones (FQs)
[edit]Quinolone antibacterial compounds were first developed in the 1960s and have been in clinical use since the 1980s.[36] FQ derivatives, such as ciprofloxacin, levofloxacin and moxifloxacin (Fig. 6) have been highly-successful. These compounds work by interacting with their target (gyrase or topo IV) and DNA at the cleavage site to stabilize the DNA-protein covalent cleavage intermediate. Specifically, they intercalate into the DNA and prevent the DNA religation step of the topoisomerase reaction (Fig. 5). This is a highly-effective mechanism of inhibition that is also used by several topoisomerase-targeted anti-cancer drugs. Despite their spectacular success, resistance to FQs is a serious problem.[36] A variety of other compounds, such as quinazolinediones and imidazolpyrazinones,[37] work in a similar manner and it is hoped that some of these will replace FQs in the future.
Aminocoumarins
[edit]Aminocoumarins (Fig. 6), such as novobiocin, clorobiocin and coumermycin A1, are natural products from Streptomyces that inhibit the ATPase reaction of gyrase and topo IV.[37] Although they can be very potent against their target, they suffer from permeability and toxicity issues, and thus have not enjoyed the level of clinical success of the FQs.
Proteinaceous inhibitors
[edit]There are a number of protein inhibitors of gyrase, including the bacterial toxins CcdB, MccB17, and ParE,[38][39][40] that stabilize the cleavage complex, in a similar manner to FQs. Although these proteins are not viable as antibacterials, their mode of action could inspire the development of novel antibacterial compounds. Other protein inhibitors of gyrase prevent DNA binding by the topoisomerase rather than stabilizing cleavage complexes. These include YacG[41] and pentapeptide repeat proteins, such as QnrB1 and MfpA;[42][43] these protein inhibitors also confer resistance to fluoroquinolones.

Anti-cancer compounds
[edit]Both human topo I and topo II (both α and β isoforms) can be targeted in anticancer chemotherapy (Fig. 7).[32][33][44][45][46][47] Most of these compounds act in a similar way to FQs, i.e. by stabilizing the DNA-protein covalent cleavage complex; for this they have become known as topoisomerase poisons, distinct from catalytic inhibitors.[33][34][48] Several human topoisomerase inhibitors are included on the World Health Organization's List of Essential Medicines.
Camptothecin (CPT)
[edit]Camptothecin (Fig. 7), originally derived from the tree Camptotheca acuminata, targets human topo I and derivatives such as topotecan and irinotecan are widely used in cancer chemotherapy.[33] Camptothecin and its derivatives act by stabilizing the topo I cleavage complex, preventing religation of the protein-mediated nick in the DNA. These interfacial inhibitors are stabilized by stacking interactions with the nicked DNA and hydrogen bonding to the enzyme. Although CPT derivatives stabilize a single-strand cleavage complex, subsequent collisions with replication or transcription machinery are thought to generate toxic double-stranded DNA breaks. These compounds are used as first or second line therapies to treat cancers including colorectal, ovarian, lung, breast, and cervical. However, CPT derivatives suffer from limitations associated with toxicity and limited therapeutic half-lives due to chemical instability. New topo I inhibitors, the indenoisoquinolines and fluoroindenoisoquinolines, overcome the limitations of CPT derivatives and are currently in clinical trials.[49]
Etoposide (VP-16)
[edit]Etoposide (Fig. 7) and its close relative teniposide (VM-26) are epipodophyllotoxin derivatives obtained from the rhizome of wild mandrake that target topo II by stabilizing the covalent cleavage complex and preventing religation of the cleaved DNA.[46] These are typically used in conjunction with other chemotherapy drugs to treat cancers including testicular tumors, small-cell lung cancer, and leukemia. Etoposide treatment can result in secondary leukemias arising from specific genomic translocations, mainly involving topo IIβ.[46]
Doxorubicin
[edit]
Doxorubicin (Fig. 7) and the related derivatives daunorubicin, epirubicin, and idarubicin are anthracyclines obtained from the bacterium Streptomyces[48] that target human topo II, stabilizing the cleavage complex in a similar manner to other topoisomerase poisons. Mitoxantrone is a synthetic anthracenedione that is chemically and functionally similar to anthracyclines.[47] The anthracyclines were the first topoisomerase inhibitors used to treat cancer and remain among the most widely employed and effective treatments for a broad range of cancers including breast cancer, lymphoma, leukemias, carcinomas, sarcomas, and other tumors.[47] These compounds are DNA intercalating agents and as such can impact a wide range of cellular DNA processes in addition to specifically poisoning topo II.[33] Additional cytotoxicity stems from redox reactions involving anthracyclines that generate reactive oxygen species. Generation of reactive oxygen, along with poisoning of topo IIβ, result in the dose-limiting cardiotoxicity of the anthracyclines.[33]
Merbarone
[edit]Merbarone is a thiobarbituric acid derivative, and dexrazoxane (ICRF-187), one of several related bisdioxopiperazine derivatives, (Fig. 7) are examples of catalytic inhibitors of topo II, i.e. they prevent completion of the catalytic cycle of topo II but do not stabilize the DNA cleavage complex. Whereas these catalytic inhibitors exhibit cytotoxicity and have been tested in clinical trials, they are not currently in clinical use for cancer therapy.[47] However, dexrazoxane, which blocks ATP hydrolysis by topo II, is used to prevent cardiotoxicity associated with the anthracyclines.[50][51]
| Topoisomerase | Subfamily type | Function | Multimericity | Metal dependence | ATP dependence | Single- or double-stranded cleavage? | Cleavage polarity | Change in link number (L) |
|---|---|---|---|---|---|---|---|---|
| Topoisomerase I
(E. coli) |
Type IA | Removes (-), but not (+) supercoils. Prevents excessive supercoiling of the genome, and supports transcription | Monomer | Yes (Mg2+) | No | SS | ||
| Topoisomerase III
(E. coli) |
Removes (-), but not (+) supercoils; overlapping function with topoisomerase IV | |||||||
| Topoisomerase IIIα
(H. sapiens) |
Removes (-), but not (+) supercoils; assists in the unlinking of precatenanes in cellular DNA replication; can catalyze the knotting, unknotting, and interlinking of single-stranded circles as well as the knotting, unknotting, catenation, and decatenation of gapped or nicked duplex DNA circles | |||||||
| Topoisomerase IIIβ
(H. sapiens) |
Has been shown to be a putative RNA topoisomerase. Involved in RNA processing | |||||||
| Reverse gyrase
(Archaea) |
Removes (-), but not (+) supercoils, introduces positive supercoils | Monomer and Heterodimer | Yes | |||||
| Topoisomerase I
(H. sapiens) |
Type IB | Removes (+) and (-) supercoils; supports fork movement during replication and transcription | Monomer | No | No | SS | 3' | ±n |
| Topoisomerase I
(Vaccinia virus) |
||||||||
| Topoisomerase V
(Archaea) |
Type IC | Relaxes (+) and (-) supercoils. Involved in DNA repair. | Monomer | No | No | SS | 3' | ±n |
| Topoisomerase II (DNA gyrase)
(E. coli) |
Type IIA | Generates (-) supercoils (the only topoisomerase known to do this) | Heterotetramer | Yes (Mg2+) | Yes | DS | 5' | ±2 |
| Topoisomerase IV
(E. coli) |
Decatenates replicated DNA; relaxes (+) supercoils faster than (-) | Heterotetramer | ||||||
| Topoisomerase IIα
(H. sapiens) |
Essential; Unlinks intertwined daughter duplexes in replication; contributes to DNA relaxation during transcription | Homodimer | ||||||
| Topoisomerase IIβ
(H. sapiens) |
Role in suppressing recombination or supporting transcription in neurons | Homodimer | ||||||
| Topoisomerase VI
(Archaea) |
Type IIB | Relaxes (+) and (-) supercoils; responsible for decatenating replication intermediates | Heterotetramer | Yes (Mg2+) | Yes | DS | 5' | ±2 |
Role of topoisomerase in transcriptional regulation
[edit]At least one topoisomerase, DNA topoisomerase II beta (topo IIβ), has a regulatory role in gene transcription. Topo IIβ–dependent double-strand DNA breaks and components of the DNA damage repair machinery are important for rapid expression of immediate early genes, as well as for signal-responsive gene regulation.[52][53][54][55] Topo IIβ, with other associated enzymes,[54] appears to be important for the release of paused RNA polymerase at highly transcribed or long genes.[56][57][58]
Topo IIβ in initiation of transcription
[edit]Stimulus-induced DNA double-strand breaks (DSBs) that are limited to a short-term (10 minutes to 2 hours) are induced by topo IIβ in the promoter regions of signal-regulated genes. These DSBs allow rapid up-regulation of expression of such signal responsive genes in a number of systems (see Table below). These signal-regulated genes include genes activated in response to stimulation with estrogen, serum, insulin, glucocorticoids (such as dexamethasone) and activation of neurons. When the induced DNA double-strand break has been repaired, then transcription of the signal-responsive gene returns to a low basal level.[52]

Topo IIβ and PARP-1 were found to be constitutively present at a moderate level near the transcription start site of a promoter of a signal-responsive gene. After the signal occurred, topo IIβ caused a double-strand break and PARP-1 was involved in replacing histone H1 by HMGB1/HMGA2, which can promote transcription.[55] Topo IIβ and PARP-1 increased at the site of the double-strand break and components of the non-homologous end joining DNA repair pathway, including DNA-PKcs, Ku70/Ku80 and DNA ligase IV assembled with topo IIβ and PARP-1. This assemblage was all present at the linker DNA adjacent to a single nucleosome in the promoter region of a gene (see Figure). The nucleosome was close to the transcription start site of the gene.[55] The components of the non-homologous end joining DNA repair pathway were essential to the closing of the DNA double-strand break.[52]
| Ligand or activating agent | Gene(s) evaluated for DSBs | DSB after signal-induced activation | DSB was required for transcription | DSB location | Proteins present at DSB | Duration of DSB after signal-induced activation | Refs |
|---|---|---|---|---|---|---|---|
| Estradiol | pS2 | yes | yes | promoter | topo IIβ, PARP-1, DNA-PKcs, Ku70, Ku80 | 10 minutes | [59] |
| Insulin | FASN | yes | yes | promoter | topo IIβ, PARP-1, DNA-PKcs, Ku70, Ku80, protein phosphatase 1 (PP1), P/CAF | 3 hours | [60] |
| Heat shock or Serum | HSPA1B, JUN, FOS, EGR1, MYC | yes | yes | promoter & POLII pausing site | DNA-PKcs, Ku70, γH2AX, TRIM28 | 30 seconds to 5 minutes | [61] |
| Dexamethasone or Estradiol | PS2, MMTV, PLZF, HSD11B2 | yes | yes | promoter | topo IIβ, PARP-1, DNA-PKcs, Ku70, Ku80, BRG1 | 15 minutes | [62] |
| KCl or NMDA activation of cultured primary cortical neurons | FOS, EGR1, NPAS4, NR4A1 | yes | yes | promoter | topo IIβ, PARP-1, DNA-PKcs, Ku70, Ku80, CTCF | Up to 2 hours | [52] |
| Fear conditioning (evaluated in mouse hippocampal and medial prefontal cortex neurons) | >200 genes with new DSBs and up-regulated expression | yes | DSBs were correlated with transcription | promoters | not tested | 10 minutes and second peak at 30 minutes | [63] |
Topo IIβ regulation of gene expression
[edit]RNA polymerase II frequently has a pausing site that is about 30–60 nucleotides downstream of the transcription start site of a gene.[64][65] The pausing of RNA polymerase II at these sites and the controlled release of the pausing is thought to have a regulatory role in gene transcription. As pointed out by Singh et al.,[58] "about 80% of highly expressed genes in HeLa cells are paused". Very short-term, but not immediately resealed, topo IIβ-induced DNA double-strand breaks occur at sites of RNA polymerase II pausing, and appear to be required for efficient release of the paused state and progression to gene transcription.[56][57][58] For the genes at which it occurs, the DNA double-stranded break induced by TOP2B is thought to be part of the process of regulation of gene expression.
See also
[edit]- DNA topology
- Supercoil
- Type I topoisomerase
- Type II topoisomerase
- Topoisomerase I
- Topoisomerase IIα
- Topoisomerase IIβ
- Topoisomerase IIIα
- Topoisomerase IIIβ
References
[edit]- ^ a b McKie SJ, Neuman KC, Maxwell A (April 2021). "DNA topoisomerases: Advances in understanding of cellular roles and multi-protein complexes via structure-function analysis". BioEssays. 43 (4): e2000286. doi:10.1002/bies.202000286. PMC 7614492. PMID 33480441. S2CID 231679533.
- ^ Sutormin DA, Galivondzhyan AK, Polkhovskiy AV, Kamalyan SO, Severinov KV, Dubiley SA (2021-03-15). "Diversity and Functions of Type II Topoisomerases". Acta Naturae. 13 (1): 59–75. doi:10.32607/actanaturae.11058. PMC 8084294. PMID 33959387.
- ^ Wang JC (February 1971). "Interaction between DNA and an Escherichia coli protein omega". Journal of Molecular Biology. 55 (3): 523–533. doi:10.1016/0022-2836(71)90334-2. PMID 4927945.
- ^ Champoux JJ, Dulbecco R (January 1972). "An activity from mammalian cells that untwists superhelical DNA--a possible swivel for DNA replication (polyoma-ethidium bromide-mouse-embryo cells-dye binding assay)". Proceedings of the National Academy of Sciences of the United States of America. 69 (1): 143–146. doi:10.1073/pnas.69.1.143. PMC 427563. PMID 4333036.
- ^ Gellert M, Mizuuchi K, O'Dea MH, Nash HA (November 1976). "DNA gyrase: an enzyme that introduces superhelical turns into DNA". Proceedings of the National Academy of Sciences of the United States of America. 73 (11): 3872–3876. Bibcode:1976PNAS...73.3872G. doi:10.1073/pnas.73.11.3872. PMC 431247. PMID 186775.
- ^ Sugino A, Peebles CL, Kreuzer KN, Cozzarelli NR (November 1977). "Mechanism of action of nalidixic acid: purification of Escherichia coli nalA gene product and its relationship to DNA gyrase and a novel nicking-closing enzyme". Proceedings of the National Academy of Sciences of the United States of America. 74 (11): 4767–4771. Bibcode:1977PNAS...74.4767S. doi:10.1073/pnas.74.11.4767. PMC 432036. PMID 200930.
- ^ Baldi MI, Benedetti P, Mattoccia E, Tocchini-Valentini GP (June 1980). "In vitro catenation and decatenation of DNA and a novel eucaryotic ATP-dependent topoisomerase". Cell. 20 (2): 461–467. doi:10.1016/0092-8674(80)90632-7. PMID 6248247. S2CID 42645648.
- ^ Liu LF, Liu CC, Alberts BM (October 1979). "T4 DNA topoisomerase: a new ATP-dependent enzyme essential for initiation of T4 bacteriophage DNA replication". Nature. 281 (5731): 456–461. Bibcode:1979Natur.281..456L. doi:10.1038/281456a0. PMID 226889. S2CID 4343962.
- ^ Stetler GL, King GJ, Huang WM (August 1979). "T4 DNA-delay proteins, required for specific DNA replication, form a complex that has ATP-dependent DNA topoisomerase activity". Proceedings of the National Academy of Sciences of the United States of America. 76 (8): 3737–3741. Bibcode:1979PNAS...76.3737S. doi:10.1073/pnas.76.8.3737. PMC 383908. PMID 226976.
- ^ a b c d Bates AD (2005). DNA topology. Anthony Maxwell (2nd ed.). Oxford: Oxford University Press. ISBN 978-0-19-154658-7. OCLC 64239232.
- ^ Watson JD, Crick FH (May 1953). "Genetical implications of the structure of deoxyribonucleic acid". Nature. 171 (4361): 964–967. Bibcode:1953Natur.171..964W. doi:10.1038/171964b0. PMID 13063483. S2CID 4256010.
- ^ Watson JD, Crick FH (April 1953). "Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid". Nature. 171 (4356): 737–738. Bibcode:1953Natur.171..737W. doi:10.1038/171737a0. PMID 13054692. S2CID 4253007.
- ^ Franklin RE, Gosling RG (July 1953). "Evidence for 2-chain helix in crystalline structure of sodium deoxyribonucleate". Nature. 172 (4369): 156–157. Bibcode:1953Natur.172..156F. doi:10.1038/172156a0. PMID 13072614. S2CID 4169572.
- ^ Cairns J (March 1963). "The bacterial chromosome and its manner of replication as seen by autoradiography". Journal of Molecular Biology. 6 (3). Elsevier: 208–213. doi:10.1016/s0022-2836(63)80070-4. PMID 14017761.
- ^ Delbrück M (September 1954). "On the Replication of Desoxyribonucleic Acid (Dna)". Proceedings of the National Academy of Sciences of the United States of America. 40 (9): 783–788. Bibcode:1954PNAS...40..783D. doi:10.1073/pnas.40.9.783. PMC 534166. PMID 16589559.
- ^ Călugăreanu G (1959). "L'intégrale de Gauss et l'analyse des nœuds tridimensionnels". Revue de Mathématiques Pure et Appliquées. 4: 5–20.
- ^ Fuller FB (April 1971). "The writhing number of a space curve". Proceedings of the National Academy of Sciences of the United States of America. 68 (4): 815–819. Bibcode:1971PNAS...68..815B. doi:10.1073/pnas.68.4.815. PMC 389050. PMID 5279522.
- ^ White JH (1969). "Self-Linking and the Gauss Integral in Higher Dimensions". American Journal of Mathematics. 91 (3): 693–728. doi:10.2307/2373348. ISSN 0002-9327. JSTOR 2373348.
- ^ Postow L, Crisona NJ, Peter BJ, Hardy CD, Cozzarelli NR (July 2001). "Topological challenges to DNA replication: conformations at the fork". Proceedings of the National Academy of Sciences of the United States of America. 98 (15): 8219–8226. Bibcode:2001PNAS...98.8219P. doi:10.1073/pnas.111006998. PMC 37424. PMID 11459956.
- ^ Sundin O, Varshavsky A (September 1981). "Arrest of segregation leads to accumulation of highly intertwined catenated dimers: dissection of the final stages of SV40 DNA replication". Cell. 25 (3): 659–669. doi:10.1016/0092-8674(81)90173-2. PMID 6269752. S2CID 24408315.
- ^ Liu LF, Wang JC (October 1987). "Supercoiling of the DNA template during transcription". Proceedings of the National Academy of Sciences of the United States of America. 84 (20): 7024–7027. Bibcode:1987PNAS...84.7024L. doi:10.1073/pnas.84.20.7024. PMC 299221. PMID 2823250.
- ^ Kraemer JA, Sanderlin AG, Laub MT (July 2019). "The Stringent Response Inhibits DNA Replication Initiation in E. coli by Modulating Supercoiling of oriC". mBio. 10 (4). doi:10.1128/mbio.01330-19. PMC 6606810. PMID 31266875.
- ^ von Freiesleben U, Rasmussen KV (September 1992). "The level of supercoiling affects the regulation of DNA replication in Escherichia coli". Research in Microbiology. 143 (7): 655–663. doi:10.1016/0923-2508(92)90060-2. PMID 1488550.
- ^ a b Kikuchi A, Asai K (1984). "Reverse gyrase--a topoisomerase which introduces positive superhelical turns into DNA". Nature. 309 (5970): 677–681. Bibcode:1984Natur.309..677K. doi:10.1038/309677a0. PMID 6328327. S2CID 4242694.
- ^ Liu LF, Liu CC, Alberts BM (March 1980). "Type II DNA topoisomerases: enzymes that can unknot a topologically knotted DNA molecule via a reversible double-strand break". Cell. 19 (3): 697–707. doi:10.1016/s0092-8674(80)80046-8. PMID 6244895. S2CID 8921868.
- ^ Brochier-Armanet C, Forterre P (May 2007). "Widespread distribution of archaeal reverse gyrase in thermophilic bacteria suggests a complex history of vertical inheritance and lateral gene transfers". Archaea. 2 (2): 83–93. doi:10.1155/2006/582916. PMC 2686386. PMID 17350929.
- ^ a b Stewart L, Redinbo MR, Qiu X, Hol WG, Champoux JJ (March 1998). "A model for the mechanism of human topoisomerase I". Science. 279 (5356): 1534–1541. Bibcode:1998Sci...279.1534S. doi:10.1126/science.279.5356.1534. PMID 9488652.
- ^ Stivers JT, Harris TK, Mildvan AS (April 1997). "Vaccinia DNA topoisomerase I: evidence supporting a free rotation mechanism for DNA supercoil relaxation". Biochemistry. 36 (17): 5212–5222. doi:10.1021/bi962880t. PMID 9136883.
- ^ Baker NM, Rajan R, Mondragón A (February 2009). "Structural studies of type I topoisomerases". Nucleic Acids Research. 37 (3): 693–701. doi:10.1093/nar/gkn1009. PMC 2647283. PMID 19106140.
- ^ Bates AD, Berger JM, Maxwell A (August 2011). "The ancestral role of ATP hydrolysis in type II topoisomerases: prevention of DNA double-strand breaks". Nucleic Acids Research. 39 (15): 6327–6339. doi:10.1093/nar/gkr258. PMC 3159449. PMID 21525132.
- ^ McKie SJ, Desai PR, Seol Y, Allen AM, Maxwell A, Neuman KC (January 2022). "Topoisomerase VI is a chirally-selective, preferential DNA decatenase". eLife. 11. doi:10.7554/eLife.67021. PMC 8837201. PMID 35076393.
- ^ a b Pommier Y (January 2013). "Drugging topoisomerases: lessons and challenges". ACS Chemical Biology. 8 (1): 82–95. doi:10.1021/cb300648v. PMC 3549721. PMID 23259582.
- ^ a b c d e f Pommier Y, Leo E, Zhang H, Marchand C (May 2010). "DNA topoisomerases and their poisoning by anticancer and antibacterial drugs". Chemistry & Biology. 17 (5): 421–433. doi:10.1016/j.chembiol.2010.04.012. PMC 7316379. PMID 20534341.
- ^ a b Pommier Y, Marchand C (December 2011). "Interfacial inhibitors: targeting macromolecular complexes". Nature Reviews. Drug Discovery. 11 (1): 25–36. doi:10.1038/nrd3404. PMC 7380715. PMID 22173432.
- ^ Tse-Dinh YC (2015). "Targeting bacterial topoisomerase I to meet the challenge of finding new antibiotics". Future Medicinal Chemistry. 7 (4): 459–471. doi:10.4155/fmc.14.157. PMC 4415981. PMID 25875873.
- ^ a b Bush NG, Diez-Santos I, Abbott LR, Maxwell A (December 2020). "Quinolones: Mechanism, Lethality and Their Contributions to Antibiotic Resistance". Molecules. 25 (23): 5662. doi:10.3390/molecules25235662. PMC 7730664. PMID 33271787.
- ^ a b Maxwell A, Bush NG, Germe T, McKie SJ (2018). "Non-quinolone topoisomerase inhibitors". In Fong IW, Drlica K (eds.). Antimicrobial resistance and implications for the twenty-first century. New York: Springer. pp. 593–618. ISBN 978-0-387-72417-1. OCLC 227210110.
- ^ Collin F, Maxwell A (August 2019). "The Microbial Toxin Microcin B17: Prospects for the Development of New Antibacterial Agents". Journal of Molecular Biology. 431 (18): 3400–3426. doi:10.1016/j.jmb.2019.05.050. PMC 6722960. PMID 31181289.
- ^ Jiang Y, Pogliano J, Helinski DR, Konieczny I (May 2002). "ParE toxin encoded by the broad-host-range plasmid RK2 is an inhibitor of Escherichia coli gyrase". Molecular Microbiology. 44 (4): 971–979. doi:10.1046/j.1365-2958.2002.02921.x. PMID 12010492. S2CID 40019620.
- ^ Smith AB, Maxwell A (2006). "A strand-passage conformation of DNA gyrase is required to allow the bacterial toxin, CcdB, to access its binding site". Nucleic Acids Research. 34 (17): 4667–4676. doi:10.1093/nar/gkl636. PMC 1635281. PMID 16963775.
- ^ Vos SM, Lyubimov AY, Hershey DM, Schoeffler AJ, Sengupta S, Nagaraja V, Berger JM (July 2014). "Direct control of type IIA topoisomerase activity by a chromosomally encoded regulatory protein". Genes & Development. 28 (13): 1485–1497. doi:10.1101/gad.241984.114. PMC 4083091. PMID 24990966.
- ^ Feng L, Mundy JE, Stevenson CE, Mitchenall LA, Lawson DM, Mi K, Maxwell A (March 2021). "The pentapeptide-repeat protein, MfpA, interacts with mycobacterial DNA gyrase as a DNA T-segment mimic". Proceedings of the National Academy of Sciences of the United States of America. 118 (11). Bibcode:2021PNAS..11816705F. doi:10.1073/pnas.2016705118. PMC 7980463. PMID 33836580.
- ^ Mazurek Ł, Ghilarov D, Michalczyk E, Pakosz Z, Metelev M, Czyszczoń W, et al. (February 2021). "Pentapeptide repeat protein QnrB1 requires ATP hydrolysis to rejuvenate poisoned gyrase complexes". Nucleic Acids Research. 49 (3): 1581–1596. doi:10.1093/nar/gkaa1266. PMC 7897471. PMID 33434265.
- ^ Nelson EM, Tewey KM, Liu LF (March 1984). "Mechanism of antitumor drug action: poisoning of mammalian DNA topoisomerase II on DNA by 4'-(9-acridinylamino)-methanesulfon-m-anisidide". Proceedings of the National Academy of Sciences of the United States of America. 81 (5): 1361–1365. Bibcode:1984PNAS...81.1361N. doi:10.1073/pnas.81.5.1361. PMC 344833. PMID 6324188.
- ^ Pommier Y, Tanizawa A, Kohn KW (1994). "Mechanisms of topoisomerase I inhibition by anticancer drugs". DNA Topoisomerases: Topoisomerase-Targeting Drugs. Advances in Pharmacology. Vol. 29B. Elsevier. pp. 73–92. doi:10.1016/s1054-3589(08)61132-1. ISBN 978-0-12-032930-4. PMID 8996602.
- ^ a b c Vann KR, Oviatt AA, Osheroff N (June 2021). "Topoisomerase II Poisons: Converting Essential Enzymes into Molecular Scissors". Biochemistry. 60 (21): 1630–1641. doi:10.1021/acs.biochem.1c00240. PMC 8209676. PMID 34008964.
- ^ a b c d Murphy MB, Mercer SL, Deweese JE (January 2017). "Inhibitors and Poisons of Mammalian Type II Topoisomerases". Advances in Molecular Toxicology. Vol. 11. Elsevier. pp. 203–240. doi:10.1016/b978-0-12-812522-9.00005-1. ISBN 9780128125229.
- ^ a b Hande KR (October 1998). "Clinical applications of anticancer drugs targeted to topoisomerase II". Biochimica et Biophysica Acta (BBA) - Gene Structure and Expression. 1400 (1–3): 173–184. doi:10.1016/s0167-4781(98)00134-1. PMID 9748560.
- ^ Pommier Y, Cushman M (May 2009). "The indenoisoquinoline noncamptothecin topoisomerase I inhibitors: update and perspectives". Molecular Cancer Therapeutics. 8 (5): 1008–1014. doi:10.1158/1535-7163.mct-08-0706. PMC 2888777. PMID 19383846.
- ^ Speyer JL, Green MD, Kramer E, Rey M, Sanger J, Ward C, et al. (September 1988). "Protective effect of the bispiperazinedione ICRF-187 against doxorubicin-induced cardiac toxicity in women with advanced breast cancer". The New England Journal of Medicine. 319 (12): 745–752. doi:10.1056/nejm198809223191203. PMID 3137469.
- ^ Cvetković RS, Scott LJ (2005). "Dexrazoxane: a review of its use for cardioprotection during anthracycline chemotherapy". Drugs. 65 (7): 1005–1024. doi:10.2165/00003495-200565070-00008. PMID 15892593.
- ^ a b c d Madabhushi R, Kim TK (March 2018). "Emerging themes in neuronal activity-dependent gene expression". Mol Cell Neurosci. 87: 27–34. doi:10.1016/j.mcn.2017.11.009. PMC 5894330. PMID 29254824.
- ^ Bunch H, Jeong J, Kang K, Jo DS, Cong AT, Kim D, Kim D, Cho DH, Lee YM, Chen BP, Schellenberg MJ, Calderwood SK (October 2021). "BRCA1-BARD1 regulates transcription through modulating topoisomerase IIβ". Open Biol. 11 (10): 210221. doi:10.1098/rsob.210221. PMC 8492178. PMID 34610268.
- ^ a b Austin CA, Cowell IG, Khazeem MM, Lok D, Ng HT (December 2021). "TOP2B's contributions to transcription". Biochem Soc Trans. 49 (6): 2483–2493. doi:10.1042/BST20200454. PMID 34747992. S2CID 243846627.
- ^ a b c Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG (June 2006). "A topoisomerase II beta-mediated dsDNA break required for regulated transcription". Science. 312 (5781): 1798–802. Bibcode:2006Sci...312.1798J. doi:10.1126/science.1127196. PMID 16794079. S2CID 206508330.
- ^ a b Dellino GI, Palluzzi F, Chiariello AM, Piccioni R, Bianco S, Furia L, De Conti G, Bouwman BA, Melloni G, Guido D, Giacò L, Luzi L, Cittaro D, Faretta M, Nicodemi M, Crosetto N, Pelicci PG (June 2019). "Release of paused RNA polymerase II at specific loci favors DNA double-strand-break formation and promotes cancer translocations". Nat Genet. 51 (6): 1011–1023. doi:10.1038/s41588-019-0421-z. PMID 31110352. S2CID 256819778.
- ^ a b Gittens WH, Johnson DJ, Allison RM, Cooper TJ, Thomas H, Neale MJ (October 2019). "A nucleotide resolution map of Top2-linked DNA breaks in the yeast and human genome". Nat Commun. 10 (1): 4846. Bibcode:2019NatCo..10.4846G. doi:10.1038/s41467-019-12802-5. PMC 6813358. PMID 31649282.
- ^ a b c Singh S, Szlachta K, Manukyan A, Raimer HM, Dinda M, Bekiranov S, Wang YH (March 2020). "Pausing sites of RNA polymerase II on actively transcribed genes are enriched in DNA double-stranded breaks". J Biol Chem. 295 (12): 3990–4000. doi:10.1074/jbc.RA119.011665. PMC 7086017. PMID 32029477.
- ^ Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG (June 2006). "A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription". Science. 312 (5781): 1798–802. Bibcode:2006Sci...312.1798J. doi:10.1126/science.1127196. PMID 16794079. S2CID 206508330.
- ^ Wong RH, Chang I, Hudak CS, Hyun S, Kwan HY, Sul HS (March 2009). "A role of DNA-PK for the metabolic gene regulation in response to insulin". Cell. 136 (6): 1056–72. doi:10.1016/j.cell.2008.12.040. PMC 2768498. PMID 19303849.
- ^ Bunch H, Lawney BP, Lin YF, Asaithamby A, Murshid A, Wang YE, Chen BP, Calderwood SK (December 2015). "Transcriptional elongation requires DNA break-induced signalling". Nat Commun. 6: 10191. Bibcode:2015NatCo...610191B. doi:10.1038/ncomms10191. PMC 4703865. PMID 26671524.
- ^ Trotter KW, King HA, Archer TK (August 2015). "Glucocorticoid Receptor Transcriptional Activation via the BRG1-Dependent Recruitment of TOP2β and Ku70/86". Mol Cell Biol. 35 (16): 2799–817. doi:10.1128/MCB.00230-15. PMC 4508321. PMID 26055322.
- ^ Stott RT, Kritsky O, Tsai LH (2021). "Profiling DNA break sites and transcriptional changes in response to contextual fear learning". PLOS ONE. 16 (7): e0249691. Bibcode:2021PLoSO..1649691S. doi:10.1371/journal.pone.0249691. PMC 8248687. PMID 34197463.
- ^ Dollinger R, Gilmour DS (July 2021). "Regulation of Promoter Proximal Pausing of RNA Polymerase II in Metazoans". J Mol Biol. 433 (14): 166897. doi:10.1016/j.jmb.2021.166897. PMC 8184617. PMID 33640324.
- ^ Price DH (May 2018). "Transient pausing by RNA polymerase II". Proc Natl Acad Sci U S A. 115 (19): 4810–4812. Bibcode:2018PNAS..115.4810P. doi:10.1073/pnas.1805129115. PMC 5949015. PMID 29691322.
Further reading
[edit]- Wang JC (2009). Untangling the Double Helix: DNA entanglement and the action of the DNA topoisomerases. Cold Spring Harbor: Cold Spring Harbor Laboratory Press. p. 245. ISBN 978-0-87969-879-9.
External links
[edit]- DNA+Topoisomerases at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
