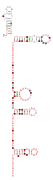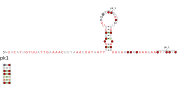RAGATH RNA motifs
| RAGATH-8 | |
|---|---|
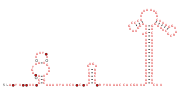
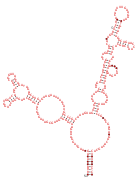
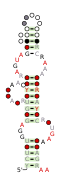
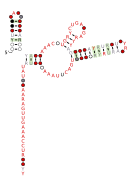
 Consensus secondary structure and sequence conservation of RAGATH-8 RNA | |
| Identifiers | |
| Symbol | RAGATH-8 |
| Rfam | RF02687 |
| Other data | |
| RNA type | Gene; sRNA |
| GO | GO:0003824 |
| SO | SO:0000370 |
| PDB structures | PDBe |
RNAs Associated with Genes Associated with Twister and Hammerhead ribozymes (RAGATH) refers to a bioinformatics strategy that was devised to find self-cleaving ribozymes in bacteria.[1] It also refers to candidate RNAs, or RAGATH RNA motifs, discovered using this strategy.
With the discovery of the twister ribozyme,[2] it was recognized that many genetic elements in bacteria are often located nearby to twister ribozymes and also to the previously discovered hammerhead ribozymes.[2] These genetic elements include several gene classes, many of which are characteristic of Mu-like phages. The nearby elements also include twister and hammerhead ribozymes. In other words, twister and hammerhead ribozymes are often located in bacteria nearby to other twister or hammerhead ribozymes.
Given these observations, researchers hypothesized that other classes of self-cleaving ribozymes would also associate with these genetic elements. Therefore, searches were conducted on the non-coding regions nearby to the associated genetic elements to find conserved RNA structures using a previously established method.[3] Such RNA structures would then be candidates as self-cleaving ribozymes.
Using this method, previously unknown self-cleaving ribozyme classes were found: the twister sister, pistol and hatchet ribozymes. Unusual examples of hammerhead and HDV ribozymes were also found. Twelve additional conserved RNA structures did not appear to function as ribozymes, and the biological and biochemical functions of these RNAs remain unknown. All conserved RNAs were named "RAGATH RNA motifs", and the unsolved RNAs are numbered RAGATH-4 through RAGATH-15. Additional RAGATH motifs that did not self-cleave 'in vitro' were also later published.[4]
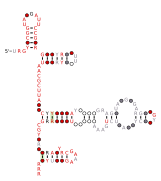
RAGATH-18 RNAs contain a predicted kink turn.[5] This particular example of a kink turn was studied to better understand how kink turn structures relate to their sequences.
References[edit]
- ^ Weinberg Z, Kim PB, Chen TH, Li S, Harris KA, Lünse CE, Breaker RR (2015). "New classes of self-cleaving ribozymes revealed by comparative genomics analysis". Nat. Chem. Biol. 11 (8): 606–10. doi:10.1038/nchembio.1846. PMC 4509812. PMID 26167874.
- ^ a b Roth A, Weinberg Z, Chen AG, Kim PB, Ames TD, Breaker RR (2014). "A widespread self-cleaving ribozyme class is revealed by bioinformatics". Nat. Chem. Biol. 10 (1): 56–60. doi:10.1038/nchembio.1386. PMC 3867598. PMID 24240507.
- ^ Weinberg Z, Wang JX, Bogue J, Yang J, Corbino K, Moy RH, Breaker RR (2010). "Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea, and their metagenomes". Genome Biol. 11 (3): R31. doi:10.1186/gb-2010-11-3-r31. PMC 2864571. PMID 20230605.
- ^ Weinberg Z, Lünse CE, Corbino KA, Ames TD, Nelson JW, Roth A, Perkins KR, Sherlock ME, Breaker RR (October 2017). "Detection of 224 candidate structured RNAs by comparative analysis of specific subsets of intergenic regions". Nucleic Acids Res. 45 (18): 10811–10823. doi:10.1093/nar/gkx699. PMC 5737381. PMID 28977401.
- ^ Huang L, Liao X, Li M, Wang J, Peng X, Wilson TJ, Lilley DM (June 2021). "Structure and folding of four putative kink turns identified in structured RNA species in a test of structural prediction rules". Nucleic Acids Res. 49 (10): 5916–5924. doi:10.1093/nar/gkab333. PMC 8191799. PMID 33978763.