Talk:DNA/Test

Deoxyribonucleic acid, or DNA is a nucleic acid molecule that contains the genetic instructions used in the development and functioning of all living organisms. The main role of DNA is the long-term storage of information and it is often compared to a set of blueprints, since DNA contains the instructions needed to construct other components of cells, such as proteins and RNA molecules. The DNA segments that carry this genetic information are called genes, but other DNA sequences have structural purposes, or are involved in regulating the use of this genetic information.
Chemically, DNA is a long polymer of simple units called nucleotides, which are held together by a backbone made of alternating sugars and phosphate groups. Attached to each sugar is one of four types of molecules called bases. It is the sequence of these four bases along the backbone that encodes information. This information is read using the genetic code, which specifies the sequence of the amino acids within proteins. The code is read by copying stretches of DNA into a complementary nucleic acid (RNA) that is structurally similar to DNA. Most of these RNA molecules are used to synthesize proteins, but others are used directly in structures such as ribosomes and spliceosomes.
Within cells, DNA is organized into structures called chromosomes and the set of chromosomes within a cell make up a genome. Eukaryotic organisms such as animals, plants, and fungi store their DNA inside the cell nucleus, while in prokaryotes such as bacteria it is found in the cell's cytoplasm. Within the chromosomes, chromatin proteins such as histones compact and organize DNA, which helps control its interactions with other proteins and control which genes are transcribed.
Physical and chemical properties[edit]
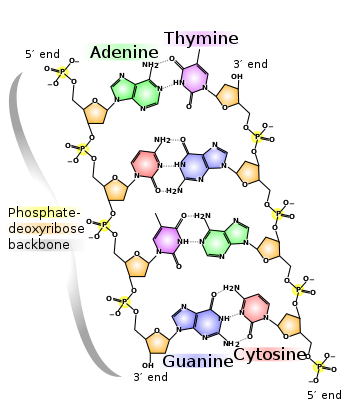
DNA is a long polymer made from repeating units called nucleotides.[1][2] The DNA chain is 22 to 24 Ångströms wide (2.2 to 2.4 nanometres), and one nucleotide unit is 3.3 Ångstroms (0.33 nanometres) long.[3] Although each individual repeating unit is very small, DNA polymers can be enormous molecules containing millions of nucleotides. For instance, the largest human chromosome, chromosome number 1, is 220 million base pairs long.[4]
In living organisms, DNA does not usually exist as a single molecule, but instead as a tightly-associated pair of molecules.[5][6] These two long strands entwine like vines, in the shape of a double helix. The nucleotide repeats contain both the backbone of the molecule, which holds the chain together, and a base, which interacts with the other DNA strand in the helix. In general, a base linked to a sugar is called a nucleoside and a base linked to a sugar and one or more phosphate groups is called a nucleotide. If multiple nucleotides are linked together, as in DNA, this polymer is referred to as a polynucleotide.[7]
The backbone of the DNA strand is made from alternating phosphate and sugar residues.[8] The sugar in DNA is 2-deoxyribose, which is a pentose (five carbon) sugar. The sugars are joined together by phosphate groups that form phosphodiester bonds between the third and fifth carbon atoms of adjacent sugar rings. These asymmetric bonds mean a strand of DNA has a direction. In a double helix the direction of the nucleotides in one strand is opposite to their direction in the other strand. This arrangement of DNA strands is called antiparallel. The asymmetric ends of a strand of DNA bases are referred to as the 5' (five prime) and 3' (three prime) ends. One of the major differences between DNA and RNA is the sugar, with 2-deoxyribose being replaced by the alternative pentose sugar ribose in RNA.[6]
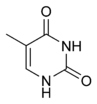
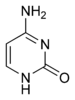
The DNA double helix is stabilized by hydrogen bonds between the bases attached to the two strands. The four bases found in DNA are adenine (abbreviated A), cytosine (C), guanine (G) and thymine (T). These four bases are shown below and are attached to the sugar/phosphate to form the complete nucleotide, as shown for adenosine monophosphate.

|

|
|
|

|
| Adenine | Guanine | Thymine | Cytosine | Adenosine monophosphate |
These bases are classified into two types; adenine and guanine are fused five- and six-membered heterocyclic compounds called purines, while cytosine and thymine are six-membered rings called pyrimidines.[7] A fifth pyrimidine base, called uracil (U), usually replaces thymine in RNA and differs from thymine by lacking a methyl group on its ring. Uracil is normally only found in DNA as a breakdown product of cytosine, but a very rare exception to this rule is a bacterial virus called PBS1 that contains uracil in its DNA.[9] In contrast, following RNA synthesis a significant number of the uracils are converted to thymines by the enzymatic addition of the missing methyl group. This occurs mostly on structural and enzymatic RNAs like transfer RNAs and ribosomal RNA.[10]

The double helix is a right-handed spiral. As the DNA strands wind around each other, they leave gaps between each set of phosphate backbones, revealing the sides of the bases inside (see animation). There are two of these grooves twisting around the surface of the double helix: one groove is 22 Å wide and the other is 12 Å wide.[12] The larger groove is called the major groove, while the smaller, narrower groove is called the minor groove. The narrowness of the minor groove means that the edges of the bases are more accessible in the major groove. As a result, proteins like transcription factors that can bind to specific sequences in double-stranded DNA usually read the sequence by making contacts to the sides of the bases exposed in the major groove.[13]

|

|
Base pairing[edit]
Each type of base on one strand forms a bond with just one type of base on the other strand. This is called complementary base pairing. Here, purines form hydrogen bonds to pyrimidines, with A bonding only to T, and C bonding only to G. This arrangement of two nucleotides joined together across the double helix is called a base pair. In a double helix, the two strands are also held together by forces generated by the hydrophobic effect and pi stacking, but these forces are not affected by the sequence of the DNA.[14] As hydrogen bonds are not covalent, they can be broken and rejoined relatively easily. The two strands of DNA in a double helix can therefore be pulled apart like a zipper, either by a mechanical force or high temperature.[15] As a result of this complementarity, all the information in the double-stranded sequence of a DNA helix is duplicated on each strand, which is vital in DNA replication. Indeed, this reversible and specific interaction between complementary base pairs is critical for all the functions of DNA in living organisms.[1]
The two types of base pairs form different numbers of hydrogen bonds, AT forming two hydrogen bonds, and GC forming three hydrogen bonds (see figures, left). The GC base pair is therefore stronger than the AT base pair. As a result, it is both the percentage of GC base pairs and the overall length of a DNA double helix that determine the strength of the association between the two strands of DNA. Long DNA helices with a high GC content have strongly interacting strands, while short helices with high AT content have weakly interacting strands.[16] Parts of the DNA double helix that need to separate easily, such as the TATAAT Pribnow box in bacterial promoters, tend to have sequences with a high AT content, making the strands easier to pull apart.[17] In the laboratory, the strength of this interaction can be measured by finding the temperature required to break the hydrogen bonds, their melting temperature (also called Tm value). When all the base pairs in a DNA double helix melt, the strands separate and exist in solution as two entirely independent molecules. These single-stranded DNA molecules have no single shape, but some conformations are more stable than others.[18]
Sense and antisense[edit]
A DNA sequence is called "sense" if its sequence is the same as that of a messenger RNA (mRNA) copy that is translated into protein. The sequence on the opposite strand is complementary to the sense sequence and is therefore called the "antisense" sequence. Since RNA polymerases work by making a complementary copy of their templates, it is this antisense strand that is the template for producing the sense mRNA. Both sense and antisense sequences can exist on different parts of the same strand of DNA. In both prokaryotes and eukaryotes, antisense sequences are transcribed, but the functions of these RNAs are not entirely clear.[19] One proposal is that antisense RNAs are involved in regulating gene expression through RNA-RNA base pairing.[20]
A few DNA sequences in prokaryotes and eukaryotes, and more in plasmids and viruses, blur the distinction made above between sense and antisense strands by having overlapping genes.[21] In these cases, some DNA sequences do double duty, encoding one protein when read 5' to 3' along one strand, and a second protein when read in the opposite direction (still 5' to 3') along the other strand. In bacteria, this overlap may be involved in the regulation of gene transcription,[22] while in viruses, overlapping genes increase the amount of information that can be encoded within the small viral genome.[23] Another way of reducing genome size is seen in some viruses that contain linear or circular single-stranded DNA as their genetic material.[24][25]
Overview of biological functions[edit]
The information carried by DNA is held in the sequence of pieces of DNA called genes. Genetic information in genes is transmitted through complementary base pairing. For example, when a cell uses the information in a gene, the DNA sequence is copied into a complementary RNA sequence in a process called transcription. Usually, this RNA copy is then used to make a matching protein sequence in a process called translation. Alternatively, a cell may simply copy its genetic information in a process called DNA replication. The details of these functions are covered in other articles; here we focus on the interactions that happen in these processes between DNA and other molecules.

DNA is located in the cell nucleus of eukaryotes, as well as small amounts in mitochondria and chloroplasts. In prokaryotes, the DNA is held within an irregularly shaped body in the cytoplasm called the nucleoid.[27] The DNA is usually in linear chromosomes in eukaryotes, and circular chromosomes in prokaryotes. In the human genome, there is approximately 3 billion base pairs of DNA arranged into 46 chromosomes.[28] The genetic information in a genome is held within genes. A gene is a unit of heredity and is a region of DNA that influences a particular characteristic in an organism. Genes contain an open reading frame that can be transcribed, as well as regulatory sequences such as promoters and enhancers, which control the expression of the open reading frame.
Transcription and translation[edit]
A gene is a sequence of DNA that contains genetic information and can influence the phenotype of an organism. Within a gene, the sequence of bases along a DNA strand defines a messenger RNA sequence, which then defines a protein sequence. The relationship between the nucleotide sequences of genes and the amino-acid sequences of proteins is determined by the rules of translation, known collectively as the genetic code. The genetic code consists of three-letter 'words' called codons formed from a sequence of three nucleotides (e.g. ACT, CAG, TTT).
In transcription, the codons of a gene are copied into messenger RNA by RNA polymerase, an enzyme that synthesises the polynucleotide chains that make up RNA from nucleoside triphosphates, the basic building blocks. This RNA copy is then decoded by a ribosome that reads the RNA sequence by base-pairing the messenger RNA to transfer RNA, which carries amino acids. Since there are 4 bases in 3-letter combinations, there are 64 possible codons ( combinations). These encode the twenty standard amino acids. Most amino acids, therefore, have more than one possible codon. There are also three 'stop' or 'nonsense' codons signifying the end of the coding region; these are the TAA, TGA and TAG codons.
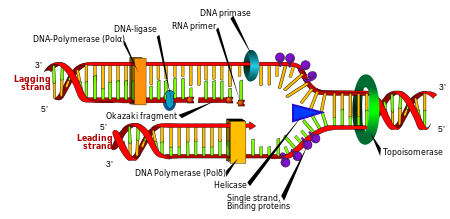
Replication[edit]
Cell division is essential for an organism to grow, but when a cell divides it must replicate the DNA in its genome so that the two daughter cells have the same genetic information as their parent. The double-stranded structure of DNA provides a simple mechanism for DNA replication. Here, the two strands are separated and then each strand's complementary DNA sequence is recreated by an enzyme called DNA polymerase. This enzyme makes the complementary strand by finding the correct base through complementary base pairing, and bonding it onto the original strand. As DNA polymerases can only extend a DNA strand in a 5' to 3' direction, different mechanisms are used to copy the antiparallel strands of the double helix.[29] In this way, the base on the old strand dictates which base appears on the new strand, and the cell ends up with a perfect copy of its DNA.
Genetic recombination[edit]

|

|
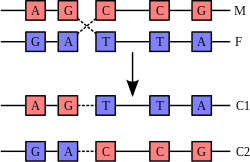
A DNA helix does not usually interact with other segments of DNA, and in human cells the different chromosomes even occupy separate areas in the nucleus called "chromosome territories".[31] This physical separation of different chromosomes is important for the ability of DNA to function as a stable repository for information, as one of the few times chromosomes interact is when they recombine. Recombination is when two DNA helices break, swap a section and then rejoin. Recombination allows chromosomes to exchange genetic information and produces new combinations of genes, which increases the efficiency of selection and can be important in the rapid evolution of new proteins.[32] Genetic recombination can also be involved in DNA repair, particularly in the cell's response to double-strand breaks.[33]
The most common form of recombination is homologous recombination, where the two chromosomes involved share very similar sequences. Non-homologous recombination can be damaging to cells, as it can produce chromosomal translocations and genetic abnormalities. The recombination reaction is catalyzed by enzymes known as recombinases, such as Cre recombinase.[34] In the first step, the recombinase creates a nick in one strand of a DNA double helix, allowing the nicked strand to pull apart from its complementary strand and anneal to one strand of the double helix on the opposite chromatid. A second nick allows the strand in the second chromatid to pull apart and anneal to the remaining strand in the first helix, forming a structure known as a cross-strand exchange or a Holliday junction. The Holliday junction is a tetrahedral junction structure that can be moved along the pair of chromosomes, swapping one strand for another. The recombination reaction is then halted by cleavage of the junction and re-ligation of the released DNA.[35]
Structure[edit]
Supercoiling[edit]
DNA can be twisted like a rope in a process called DNA supercoiling. Normally, with DNA in its "relaxed" state, a strand circles the axis of the double helix once every 10.4 base pairs, but if the DNA is twisted the strands become more tightly or more loosely wound.[36] If the DNA is twisted in the direction of the helix, this is positive supercoiling, and the bases are held more tightly together. If they are twisted in the opposite direction, this is negative supercoiling, and the bases come apart more easily. In nature, most DNA has slight negative supercoiling that is introduced by enzymes called topoisomerases.[37] These enzymes are also needed to relieve the twisting stresses introduced into DNA strands during processes such as transcription and DNA replication.[38]

Alternative double-helical structures[edit]
DNA exists in several possible conformations. The conformations so far identified are: A-DNA, B-DNA, C-DNA, D-DNA,[39] E-DNA,[40] H-DNA,[41] L-DNA,[39], P-DNA[42], and Z-DNA.[8][43] However, only A-DNA, B-DNA, and Z-DNA have been observed in naturally occurring biological systems. Which conformation DNA adopts depends on the sequence of the DNA, the amount and direction of supercoiling, chemical modifications of the bases and also solution conditions, such as the concentration of metal ions and polyamines.[44] Of these three conformations, the "B" form described above is most common under the conditions found in cells. The two alternative double-helical forms of DNA differ in their geometry and dimensions.
The A form is a wider right-handed spiral, with a shallow and wide minor groove and a narrower and deeper major groove. The A form occurs under non-physiological conditions in dehydrated samples of DNA, while in the cell it may be produced in hybrid pairings of DNA and RNA strands.[45] Segments of DNA where the bases have been methylated may undergo a larger change in conformation and adopt the Z form. Here, the strands turn about the helical axis in a left-handed spiral, the opposite of the more common B form.[46]

Quadruplex structures[edit]
At the ends of the linear chromosomes are specialized regions of DNA called telomeres. The main function of these regions is to allow the cell to replicate chromosome ends using the enzyme telomerase, as normal DNA polymerases working on the lagging strand cannot copy the extreme 3' ends of their DNA templates.[48] If a chromosome lacked telomeres it would become shorter each time it was replicated. These specialized chromosome caps also help protect the DNA ends from exonucleases and stop the DNA repair systems in the cell from treating them as damage to be corrected.[49] In human cells, telomeres are usually lengths of single-stranded DNA containing several thousand repeats of a simple TTAGGG sequence.[50]
These guanine-rich sequences may stabilize chromosome ends by forming very unusual quadruplex structures. Here, four guanine bases form a flat plate, through hydrogen bonding, and these flat four-base units then stack on top of each other, to form a stable quadruplex.[51] These structures are often stabilized by chelation of a metal ion in the centre of each four-base unit. The structure shown to the left is of a quadruplex formed by a DNA sequence containing four consecutive human telomere repeats. The single DNA strand forms a loop, with the sets of four bases stacking in a central quadruplex three plates deep. In the space at the centre of the stacked bases are three chelated potassium ions.[52] Other structures can also be formed and the central set of four bases can come from either one folded strand, or several different parallel strands.
In addition to these stacked structures, telomeres also form large loop structures called telomere loops, or T-loops. Here, the single-stranded DNA curls around in a circle stabilized by telomere-binding proteins.[53] The very end of the T-loop, the single-stranded telomere DNA is held onto a region of double-stranded DNA by the telomere strand disrupting the double-helical DNA and base pairing to one of the two strands. This triple-stranded structure is called a displacement loop or D-loop.[51]
Chemical modifications[edit]
Regulatory base modifications[edit]
The expression of genes is influenced by modifications of the bases in DNA. In humans, the most common base modification is cytosine methylation to produce 5-methylcytosine. This modification reduces gene expression and is important in X-chromosome inactivation.[54] The level of methylation varies between organisms, with Caenorhabditis elegans lacking cytosine methylation, while vertebrates show high levels, with up to 1% of their DNA being 5-methylcytosine.[55] Unfortunately, the spontaneous deamination of 5-methylcytosine produces thymine, and methylated cytosines are therefore mutation hotspots.[56] Other base modifications include adenine methylation in bacteria and the glycosylation of uracil to produce the "J-base" in kinetoplastids.[57][58]
DNA damage[edit]

DNA can be damaged by many different sorts of mutagens. These include oxidizing agents, alkylating agents and also high-energy electromagnetic radiation such as ultraviolet light and x-rays. The type of DNA damage produced depends on the type of mutagen. For example, UV light mostly damages DNA by producing thymine dimers, which are cross-links between adjacent pyrimidine bases in a DNA strand.[60] On the other hand, oxidants such as free radicals or hydrogen peroxide produce multiple forms of damage, including base modifications, particularly of guanosine, as well as double-strand breaks.[61] It has been estimated that in each human cell, about 500 bases suffer oxidative damage per day.[62][63] Of these oxidative lesions, the most dangerous are double-strand breaks, as these lesions are difficult to repair and can produce point mutations, insertions and deletions from the DNA sequence, as well as chromosomal translocations.[64]
Many mutagens intercalate into the space between two adjacent base pairs. Intercalators are mostly polycyclic, aromatic, and planar molecules, and include ethidium, proflavin, daunomycin, doxorubicin and thalidomide. DNA intercalators are used in chemotherapy to inhibit DNA replication in rapidly-growing cancer cells.[65] In order for an intercalator to fit between base pairs, the bases must separate, distorting the DNA strands by unwinding of the double helix. These structural modifications inhibit transcription and replication processes, causing both toxicity and mutations. As a result, DNA intercalators are often carcinogens, with benzopyrene diol epoxide, acridines, aflatoxin and ethidium bromide being well-known examples.[66][67][68]
Interactions with proteins[edit]
All the functions of DNA depend on interactions with proteins. These protein interactions can either be non-specific or specific, where the protein can only bind to a particular DNA sequence. Enzymes can also bind to DNA and of these, the polymerases that copy the DNA base sequence in transcription and DNA replication are particularly important.
DNA-binding proteins[edit]
|
| File:Nucleosome (opposites attracts).JPG |
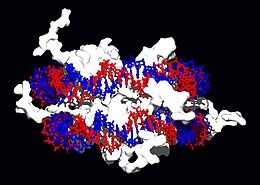
Structural proteins that bind DNA are well-understood examples of non-specific DNA-protein interactions. Within chromosomes, DNA is held in complexes between DNA and structural proteins. These proteins organize the DNA into a compact structure called chromatin. In eukaryotes this structure involves DNA binding to a complex of small basic proteins called histones, while in prokaryotes multiple types of proteins are involved.[69] The histones form a disk-shaped complex called a nucleosome, which contains two complete turns of double-stranded DNA wrapped around its surface. These non-specific interactions are formed through basic residues in the histones making ionic bonds to the acidic sugar-phosphate backbone of the DNA, and are therefore largely independent of the base sequence.[70] Chemical modifications of these basic amino acid residues include methylation, phosphorylation and acetylation.[71] These chemical changes alter the strength of the interaction between the DNA and the histones, making the DNA more or less accessible to transcription factors and changing the rate of transcription.[72] Other non-specific DNA-binding proteins found in chromatin include the high-mobility group proteins, which bind preferentially to bent or distorted DNA.[73] These proteins are important in bending arrays of nucleosomes and arranging them into more complex chromatin structures.[74]
A distinct group of DNA-binding proteins are the single-stranded DNA-binding proteins that specifically bind single-stranded DNA. In humans, replication protein A is the best-characterised member of this family and is essential for most processes where the double helix is separated, including DNA replication, recombination and DNA repair.[75] These binding proteins seem to stabilize single-stranded DNA and protect it from forming stem loops or being degraded by nucleases.

In contrast, other proteins have evolved to specifically bind particular DNA sequences. The most intensively studied of these are the various classes of transcription factors. These proteins control gene transcription. Each one of these proteins bind to one particular set of DNA sequences and thereby activates or inhibits the transcription of genes with these sequences close to their promoters. The transcription factors do this in two ways. Firstly, they can bind the RNA polymerase responsible for transcription, either directly or through other mediator proteins; this locates the polymerase at the promoter and allows it to begin transcription.[77] Alternatively, transcription factors can bind enzymes that modify the histones at the promoter; this will change the accessibility of the DNA template to the polymerase.[78]
As these DNA targets can occur throughout an organism's genome, changes in the activity of one type of transcription factor can affect thousands of genes.[79] Consequently, these proteins are often the targets of the signal transduction processes that mediate responses to environmental changes or cellular differentiation and development. The specificity of these transcription factors' interactions with DNA come from the proteins making multiple contacts to the edges of the DNA bases, allowing them to "read" the DNA sequence. Most of these base interactions are made in the major groove, where the bases are most accessible.[80]

DNA-modifying enzymes[edit]
Nucleases and ligases[edit]
Nucleases are enzymes that cut DNA strands by catalyzing the hydrolysis of the phosphodiester bonds. Nucleases that hydrolyse nucleotides from the ends of DNA strands are called exonucleases, while endonucleases cut within strands. The most frequently-used nucleases in molecular biology are the restriction endonucleases, which cut DNA at specific sequences. For instance, the EcoRV enzyme shown to the left recognizes the 6-base sequence 5'-GAT|ATC-3' and makes a cut at the vertical line. In nature, these enzymes protect bacteria against phage infection by digesting the phage DNA when it enters the bacterial cell, acting as part of the restriction modification system.[82] In technology, these sequence-specific nucleases are used in molecular cloning and DNA fingerprinting.
Enzymes called DNA ligases can rejoin cut or broken DNA strands, using the energy from either adenosine triphosphate or nicotinamide adenine dinucleotide.[83] Ligases are particularly important in lagging strand DNA replication, as they join together the short segments of DNA produced at the replication fork into a complete copy of the DNA template. They are also used in DNA repair and genetic recombination.[83]
Topoisomerases and helicases[edit]
Topoisomerases are enzymes with both nuclease and ligase activity. These proteins change the amount of supercoiling in DNA. Some of these enzyme work by cutting the DNA helix and allowing one section to rotate, thereby reducing its level of supercoiling; the enzyme then seals the DNA break.[37] Other types of these enzymes are capable of cutting one DNA helix and then passing a second strand of DNA through this break, before rejoining the helix.[84] Topoisomerases are required for many processes involving DNA, such as DNA replication and transcription.[38]
Helicases are proteins that are a type of molecular motor. They use the chemical energy in nucleoside triphosphates, predominantly ATP, to break hydrogen bonds between bases and unwind the DNA double helix into single strands.[85] These enzymes are essential for most processes where enzymes need to access the DNA bases.
Polymerases[edit]
Polymerases are enzymes that synthesise polynucleotide chains from nucleoside triphosphates. They function by adding nucleotides onto the 3ˈ hydroxyl group of the previous nucleotide in the DNA strand. As a consequence, all polymerases work in a 5' to 3' direction.[86] In the active site of these enzymes, the nucleoside triphosphate substrate base-pairs to a single-stranded polynucleotide template: this allows polymerases to accurately synthesise the complementary strand of this template. Polymerases are classified depending of the type of template they use.
In DNA replication, a DNA-dependent DNA polymerase makes a DNA copy of a DNA sequence. Accuracy is vital in this process, so many of these polymerases have a proofreading activity. Here, the polymerase recognizes the occasional mistakes in the synthesis reaction by the lack of base pairing between the mismatched nucleotides. If a mismatch is detected, a 3' to 5' exonuclease activity is activated and the incorrect base removed.[87] In most organisms DNA polymerases function in a large complex called the replisome that contains multiple accessory subunits, such as the DNA clamp or helicases.[88]
RNA-dependent DNA polymerases are a specialised class of polymerases that copy the sequence of a RNA strand into DNA. They include reverse transcriptase, which is a viral enzyme involved in the infection of cells by retroviruses, and telomerase, which is required for the replication of telomeres.[89][48] Telomerase is an unusual polymerase because it contains its own RNA template as part of its structure.[49]
Transcription is carried out by a DNA-dependent RNA polymerase that copies the sequence of a DNA strand into RNA. To begin transcribing a gene, the RNA polymerase binds to a sequence of DNA called a promoter and separates the DNA strands. It then copies the gene sequence into a messenger RNA transcript until it reaches a region of DNA called the terminator, where it halts and detaches from the DNA. As with human DNA-dependent DNA polymerases, RNA polymerase II, the enzyme that transcribes most of the genes in the human genome, operates as part of a large protein complex with multiple regulatory and accessory subunits.[90]
Evolution of DNA-based metabolism[edit]
DNA contains the genetic information that allows all modern living things to function, grow and reproduce. However, it is unclear how long in the 4-billion-year history of life DNA has performed this function, as it has been proposed that the earliest forms of life may have used RNA as their genetic material.[86][91] RNA may have acted as the central part of early cell metabolism as it can both transmit genetic information and carry out catalysis as part of ribozymes.[92] This ancient RNA world where nucleic acid would have been used for both catalysis and genetics may have influenced the evolution of the current genetic code based on four nucleotide bases. This would occur since the number of unique bases in such an organism is a trade-off between a small number of bases increasing replication accuracy and a large number of bases increasing the catalytic efficiency of ribozymes.[93]
Unfortunately, there is no direct evidence of ancient genetic systems, as recovery of DNA from most fossils is impossible. This is because DNA will survive in the environment for less than one million years and slowly degrades into short fragments in solution.[94] Although claims for older DNA have been made, most notably a report of the isolation of a viable bacterium from a salt crystal 250-million years old,[95] these claims are controversial and have been disputed.[96][97]
History[edit]

DNA was first isolated by Friedrich Miescher who, in 1869, discovered a microscopic substance in the pus of discarded surgical bandages. As it resided in the nuclei of cells, he called it "nuclein".[98] In 1929 this discovery was followed by Phoebus Levene's identification of the base, sugar and phosphate nucleotide unit.[99] Levene suggested that DNA consisted of a string of nucleotide units linked together through the phosphate groups. However, Levene thought the chain was short and the bases repeated in a fixed order. In 1937 William Astbury produced the first X-ray diffraction patterns that showed that DNA had a regular structure.[100]
In 1943, Oswald Theodore Avery discovered that traits of the "smooth" form of the Pneumococcus could be transferred to the "rough" form of the same bacteria by mixing killed "smooth" bacteria with the live "rough" form. Avery identified DNA as this transforming principle.[101] DNA's role in heredity was confirmed in 1953, when Alfred Hershey and Martha Chase in the Hershey-Chase experiment showed that DNA is the genetic material of the T2 phage.[102]
In 1953, based on X-ray diffraction images[103] taken by Rosalind Franklin and the information that the bases were paired, James D. Watson and Francis Crick suggested[103] what is now accepted as the first accurate model of DNA structure in the journal Nature.[5] Experimental evidence for Watson and Crick's model were published in a series of five articles in the same issue of Nature.[104] Of these, Franklin and Raymond Gosling's paper[105] saw the publication of the X-ray diffraction image, which was key in Watson and Crick interpretation, as well as another article, co-authored by Maurice Wilkins and his colleagues.[106] Franklin and Gosling's subsequent paper identified the distinctions between the A and B structures of the double helix in DNA.[107] In 1962 Watson, Crick, and Maurice Wilkins jointly received the Nobel Prize in Physiology or Medicine (Franklin didn't share the prize with them since she had died earlier).[108]
In an influential presentation in 1957, Crick laid out the "Central Dogma" of molecular biology, which foretold the relationship between DNA, RNA, and proteins, and articulated the "adaptor hypothesis".[109] Final confirmation of the replication mechanism that was implied by the double-helical structure followed in 1958 through the Meselson-Stahl experiment.[110] Further work by Crick and coworkers showed that the genetic code was based on non-overlapping triplets of bases, called codons, allowing Har Gobind Khorana, Robert W. Holley and Marshall Warren Nirenberg to decipher the genetic code.[111] These findings represent the birth of molecular biology.
Uses in technology[edit]
Modern biology and biochemistry make intensive use of recombinant DNA technology. Recombinant DNA is a man-made DNA sequence assembled from other DNA sequences in a plasmid. These plasmids can be transformed into organisms. The genetically modified organisms produced can be used to produce products such as recombinant proteins, used in medical research,[112] or be grown in agriculture.[113][114]
Forensics[edit]
Forensic scientists can use DNA in blood, semen, skin, saliva or hair at a crime scene to identify a perpetrator. This process is called genetic fingerprinting, or more accurately, DNA profiling. In DNA profiling, the lengths of variable sections of repetitive DNA, such as short tandem repeats and minisatellites, are compared between people. This method is usually an extremely reliable technique for identifying a criminal.[115] However, identification can be complicated if the scene is contaminated with DNA from several people.[116] DNA profiling was developed in 1984 by British geneticist Sir Alec Jeffreys,[117] and first used in forensic science to convict Colin Pitchfork in the 1988 Enderby murders case.[118] People convicted of certain types of crimes may be required to provide a sample of DNA for a database. This has helped investigators solve old cases where only a DNA sample was obtained from the scene. DNA profiling can also be used to identify victims of mass casualty incidents.[119]
Bioinformatics[edit]
Bioinformatics involves the manipulation, searching, and data mining of DNA sequence data. The development of techniques to store and search DNA sequences have led to widely-applied advances in computer science, especially string searching algorithms, machine learning and database theory.[120] String searching or matching algorithms, which find an occurrence of a sequence of letters inside a larger sequence of letters, were developed to search for specific sequences of nucleotides.[121] In other applications such as text editors, even simple algorithms for this problem usually suffice, but DNA sequences cause these algorithms to exhibit near-worst-case behaviour due to their small number of distinct characters. The related problem of sequence alignment aims to identify homologous sequences and locate the specific mutations that make them distinct. These techniques, especially multiple sequence alignment, are used in studying phylogenetic relationships and protein function.[122] Data sets representing entire genomes' worth of DNA sequences, such as those produced by the Human Genome Project, are difficult to use without annotations, which label the locations of genes and regulatory elements on each chromosome. Regions of DNA sequence that have the characteristic patterns associated with protein- or RNA-coding genes can be identified by gene finding algorithms, which allow researchers to predict the presence of particular gene products in an organism even before they have been isolated experimentally.[123]
DNA and computation[edit]
DNA was first used in computing to solve a small version of the directed Hamiltonian path problem, an NP-complete problem.[124] DNA computing is advantageous over electronic computers in power use, space use, and efficiency, due to its ability to compute in a highly parallel fashion (see parallel computing). A number of other problems, including simulation of various abstract machines, the boolean satisfiability problem, and the bounded version of the travelling salesman problem, have since been analysed using DNA computing.[125] Due to its compactness, DNA also has a theoretical role in cryptography, where in particular it allows unbreakable one-time pads to be efficiently constructed and used.[126]
History and anthropology[edit]
Because DNA collects mutations over time, which are then inherited, it contains historical information and by comparing DNA sequences, geneticists can infer the evolutionary history of organisms, their phylogeny.[127] This field of phylogenetics is a powerful tool in evolutionary biology. If DNA sequences within a species are compared, population geneticists can learn the history of particular populations. This can be used in studies ranging from ecological genetics to anthropology; for example, DNA evidence is being used to try to identify the Ten Lost Tribes of Israel.[128][129]
DNA has also been used to look at modern family relationships, such as establishing family relationships between the descendants of Sally Hemings and Thomas Jefferson. This usage is closely related to the use of DNA in criminal investigations detailed above. Indeed, some criminal investigations have been solved when DNA from crime scenes has matched relatives of the guilty individual.[130]
See also[edit]
- Genetic disorder
- Plasmid
- DNA sequencing
- Southern blot
- DNA microarray
- Polymerase chain reaction
- Protein-DNA interaction site predictor
- Phosphoramidite
- Quantification of nucleic acids
References[edit]
- ^ a b Alberts, Bruce (2002). Molecular Biology of the Cell; Fourth Edition. New York and London: Garland Science. ISBN 0-8153-3218-1.
{{cite book}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Butler, John M. (2001) Forensic DNA Typing "Elsevier". pp. 14 – 15. ISBN 978-0-12-147951-0.
- ^ Mandelkern M, Elias J, Eden D, Crothers D (1981). "The dimensions of DNA in solution". J Mol Biol. 152 (1): 153–61. PMID 7338906.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Gregory S; et al. (2006). "The DNA sequence and biological annotation of human chromosome 1". Nature. 441 (7091): 315–21. PMID 16710414.
{{cite journal}}: Explicit use of et al. in:|author=(help) - ^ a b Watson J, Crick F (1953). "Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid" (PDF). Nature. 171 (4356): 737–8. PMID 13054692.
- ^ a b Berg J., Tymoczko J. and Stryer L. (2002) Biochemistry. W. H. Freeman and Company ISBN 0-7167-4955-6
- ^ a b Abbreviations and Symbols for Nucleic Acids, Polynucleotides and their Constituents IUPAC-IUB Commission on Biochemical Nomenclature (CBN) Accessed 03 Jan 2006
- ^ a b Ghosh A, Bansal M (2003). "A glossary of DNA structures from A to Z". Acta Crystallogr D Biol Crystallogr. 59 (Pt 4): 620–6. PMID 12657780.
- ^ Takahashi I, Marmur J. (1963). "Replacement of thymidylic acid by deoxyuridylic acid in the deoxyribonucleic acid of a transducing phage for Bacillus subtilis". Nature. 197: 794–5. PMID 13980287.
- ^ Agris P (2004). "Decoding the genome: a modified view". Nucleic Acids Res. 32 (1): 223–38. PMID 14715921.
- ^ Created from PDB 1D65
- ^ Wing R, Drew H, Takano T, Broka C, Tanaka S, Itakura K, Dickerson R (1980). "Crystal structure analysis of a complete turn of B-DNA". Nature. 287 (5784): 755–8. PMID 7432492.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Pabo C, Sauer R. "Protein-DNA recognition". Annu Rev Biochem. 53: 293–321. PMID 6236744.
- ^ Ponnuswamy P, Gromiha M (1994). "On the conformational stability of oligonucleotide duplexes and tRNA molecules". J Theor Biol. 169 (4): 419–32. PMID 7526075.
- ^ Clausen-Schaumann H, Rief M, Tolksdorf C, Gaub H (2000). "Mechanical stability of single DNA molecules". Biophys J. 78 (4): 1997–2007. PMID 10733978.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Chalikian T, Völker J, Plum G, Breslauer K (1999). "A more unified picture for the thermodynamics of nucleic acid duplex melting: a characterization by calorimetric and volumetric techniques". Proc Natl Acad Sci U S A. 96 (14): 7853–8. PMID 10393911.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ deHaseth P, Helmann J (1995). "Open complex formation by Escherichia coli RNA polymerase: the mechanism of polymerase-induced strand separation of double helical DNA". Mol Microbiol. 16 (5): 817–24. PMID 7476180.
- ^ Isaksson J, Acharya S, Barman J, Cheruku P, Chattopadhyaya J (2004). "Single-stranded adenine-rich DNA and RNA retain structural characteristics of their respective double-stranded conformations and show directional differences in stacking pattern". Biochemistry. 43 (51): 15996–6010. PMID 15609994.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Hüttenhofer A, Schattner P, Polacek N (2005). "Non-coding RNAs: hope or hype?". Trends Genet. 21 (5): 289–97. PMID 15851066.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Munroe S (2004). "Diversity of antisense regulation in eukaryotes: multiple mechanisms, emerging patterns". J Cell Biochem. 93 (4): 664–71. PMID 15389973.
- ^ Makalowska I, Lin C, Makalowski W (2005). "Overlapping genes in vertebrate genomes". Comput Biol Chem. 29 (1): 1–12. PMID 15680581.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Johnson Z, Chisholm S (2004). "Properties of overlapping genes are conserved across microbial genomes". Genome Res. 14 (11): 2268–72. PMID 15520290.
- ^ Lamb R, Horvath C (1991). "Diversity of coding strategies in influenza viruses". Trends Genet. 7 (8): 261–6. PMID 1771674.
- ^ Davies J, Stanley J (1989). "Geminivirus genes and vectors". Trends Genet. 5 (3): 77–81. PMID 2660364.
- ^ Berns K (1990). "Parvovirus replication". Microbiol Rev. 54 (3): 316–29. PMID 2215424.
- ^ Created from PDB 1MSW
- ^ Thanbichler M, Wang S, Shapiro L (2005). "The bacterial nucleoid: a highly organized and dynamic structure". J Cell Biochem. 96 (3): 506–21. PMID 15988757.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Venter J; et al. (2001). "The sequence of the human genome". Science. 291 (5507): 1304–51. PMID 11181995.
{{cite journal}}: Explicit use of et al. in:|author=(help) - ^ Albà M (2001). "Replicative DNA polymerases". Genome Biol. 2 (1): REVIEWS3002. PMID 11178285.
- ^ Created from PDB 1M6G
- ^ Cremer T, Cremer C (2001). "Chromosome territories, nuclear architecture and gene regulation in mammalian cells". Nat Rev Genet. 2 (4): 292–301. PMID 11283701.
- ^ Pál C, Papp B, Lercher M (2006). "An integrated view of protein evolution". Nat Rev Genet. 7 (5): 337–48. PMID 16619049.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ O'Driscoll M, Jeggo P (2006). "The role of double-strand break repair - insights from human genetics". Nat Rev Genet. 7 (1): 45–54. PMID 16369571.
- ^ Ghosh K, Van Duyne G (2002). "Cre-loxP biochemistry". Methods. 28 (3): 374–83. PMID 12431441.
- ^ Dickman M, Ingleston S, Sedelnikova S, Rafferty J, Lloyd R, Grasby J, Hornby D (2002). "The RuvABC resolvasome". Eur J Biochem. 269 (22): 5492–501. PMID 12423347.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Benham C, Mielke S. "DNA mechanics". Annu Rev Biomed Eng. 7: 21–53. PMID 16004565.
- ^ a b Champoux J. "DNA topoisomerases: structure, function, and mechanism". Annu Rev Biochem. 70: 369–413. PMID 11395412.
- ^ a b Wang J (2002). "Cellular roles of DNA topoisomerases: a molecular perspective". Nat Rev Mol Cell Biol. 3 (6): 430–40. PMID 12042765.
- ^ a b Hayashi G, Hagihara M, Nakatani K (2005). "Application of L-DNA as a molecular tag". Nucleic Acids Symp Ser (Oxf). 49: 261–262. PMID 17150733.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Vargason JM, Eichman BF, Ho PS (2000). "The extended and eccentric E-DNA structure induced by cytosine methylation or bromination". Nature Structural Biology. 7: 758–761. PMID 10966645.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Wang G, Vasquez KM (2006). "Non-B DNA structure-induced genetic instability". Mutat Res. 598 (1–2): 103–119. PMID 16516932.
- ^ Allemand; et al. (1998). "Stretched and overwound DNA forms a Pauling-like structure with exposed bases". PNAS. 24: 14152–14157. PMID 9826669.
{{cite journal}}: Explicit use of et al. in:|author=(help) - ^ Palecek E (1991). "Local supercoil-stabilized DNA structures". Critical Reviews in Biochemistry and Molecular Biology. 26 (2): 151–226. PMID 1914495.
- ^ Basu H, Feuerstein B, Zarling D, Shafer R, Marton L (1988). "Recognition of Z-RNA and Z-DNA determinants by polyamines in solution: experimental and theoretical studies". J Biomol Struct Dyn. 6 (2): 299–309. PMID 2482766.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Wahl M, Sundaralingam M (1997). "Crystal structures of A-DNA duplexes". Biopolymers. 44 (1): 45–63. PMID 9097733.
- ^ Rothenburg S, Koch-Nolte F, Haag F. "DNA methylation and Z-DNA formation as mediators of quantitative differences in the expression of alleles". Immunol Rev. 184: 286–98. PMID 12086319.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Created from NDB UD0017
- ^ a b Greider C, Blackburn E (1985). "Identification of a specific telomere terminal transferase activity in Tetrahymena extracts". Cell. 43 (2 Pt 1): 405–13. PMID 3907856.
- ^ a b Nugent C, Lundblad V (1998). "The telomerase reverse transcriptase: components and regulation". Genes Dev. 12 (8): 1073–85. PMID 9553037.
- ^ Wright W, Tesmer V, Huffman K, Levene S, Shay J (1997). "Normal human chromosomes have long G-rich telomeric overhangs at one end". Genes Dev. 11 (21): 2801–9. PMID 9353250.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b Burge S, Parkinson G, Hazel P, Todd A, Neidle S (2006). "Quadruplex DNA: sequence, topology and structure". Nucleic Acids Res. 34 (19): 5402–15. PMID 17012276.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Parkinson G, Lee M, Neidle S (2002). "Crystal structure of parallel quadruplexes from human telomeric DNA". Nature. 417 (6891): 876–80. PMID 12050675.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Griffith J, Comeau L, Rosenfield S, Stansel R, Bianchi A, Moss H, de Lange T (1999). "Mammalian telomeres end in a large duplex loop". Cell. 97 (4): 503–14. PMID 10338214.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Klose R, Bird A (2006). "Genomic DNA methylation: the mark and its mediators". Trends Biochem Sci. 31 (2): 89–97. PMID 16403636.
- ^ Bird A (2002). "DNA methylation patterns and epigenetic memory". Genes Dev. 16 (1): 6–21. PMID 11782440.
- ^ Walsh C, Xu G. "Cytosine methylation and DNA repair". Curr Top Microbiol Immunol. 301: 283–315. PMID 16570853.
- ^ Ratel D, Ravanat J, Berger F, Wion D (2006). "N6-methyladenine: the other methylated base of DNA". Bioessays. 28 (3): 309–15. PMID 16479578.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Gommers-Ampt J, Van Leeuwen F, de Beer A, Vliegenthart J, Dizdaroglu M, Kowalak J, Crain P, Borst P (1993). "beta-D-glucosyl-hydroxymethyluracil: a novel modified base present in the DNA of the parasitic protozoan T. brucei". Cell. 75 (6): 1129–36. PMID 8261512.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Created from PDB 1JDG
- ^ Douki T, Reynaud-Angelin A, Cadet J, Sage E (2003). "Bipyrimidine photoproducts rather than oxidative lesions are the main type of DNA damage involved in the genotoxic effect of solar UVA radiation". Biochemistry. 42 (30): 9221–6. PMID 12885257.
{{cite journal}}: CS1 maint: multiple names: authors list (link), - ^ Cadet J, Delatour T, Douki T, Gasparutto D, Pouget J, Ravanat J, Sauvaigo S (1999). "Hydroxyl radicals and DNA base damage". Mutat Res. 424 (1–2): 9–21. PMID 10064846.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Shigenaga M, Gimeno C, Ames B (1989). "Urinary 8-hydroxy-2'-deoxyguanosine as a biological marker of in vivo oxidative DNA damage". Proc Natl Acad Sci U S A. 86 (24): 9697–701. PMID 2602371.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Cathcart R, Schwiers E, Saul R, Ames B (1984). "Thymine glycol and thymidine glycol in human and rat urine: a possible assay for oxidative DNA damage" (PDF). Proc Natl Acad Sci U S A. 81 (18): 5633–7. PMID 6592579.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Valerie K, Povirk L (2003). "Regulation and mechanisms of mammalian double-strand break repair". Oncogene. 22 (37): 5792–812. PMID 12947387.
- ^ Braña M, Cacho M, Gradillas A, de Pascual-Teresa B, Ramos A (2001). "Intercalators as anticancer drugs". Curr Pharm Des. 7 (17): 1745–80. PMID 11562309.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Ferguson L, Denny W (1991). "The genetic toxicology of acridines". Mutat Res. 258 (2): 123–60. PMID 1881402.
- ^ Jeffrey A (1985). "DNA modification by chemical carcinogens". Pharmacol Ther. 28 (2): 237–72. PMID 3936066.
- ^ Stephens T, Bunde C, Fillmore B (2000). "Mechanism of action in thalidomide teratogenesis". Biochem Pharmacol. 59 (12): 1489–99. PMID 10799645.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Sandman K, Pereira S, Reeve J (1998). "Diversity of prokaryotic chromosomal proteins and the origin of the nucleosome". Cell Mol Life Sci. 54 (12): 1350–64. PMID 9893710.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Luger K, Mäder A, Richmond R, Sargent D, Richmond T (1997). "Crystal structure of the nucleosome core particle at 2.8 A resolution". Nature. 389 (6648): 251–60. PMID 9305837.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Jenuwein T, Allis C (2001). "Translating the histone code". Science. 293 (5532): 1074–80. PMID 11498575.
- ^ Ito T. "Nucleosome assembly and remodelling". Curr Top Microbiol Immunol. 274: 1–22. PMID 12596902.
- ^ Thomas J (2001). "HMG1 and 2: architectural DNA-binding proteins". Biochem Soc Trans. 29 (Pt 4): 395–401. PMID 11497996.
- ^ Grosschedl R, Giese K, Pagel J (1994). "HMG domain proteins: architectural elements in the assembly of nucleoprotein structures". Trends Genet. 10 (3): 94–100. PMID 8178371.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Iftode C, Daniely Y, Borowiec J (1999). "Replication protein A (RPA): the eukaryotic SSB". Crit Rev Biochem Mol Biol. 34 (3): 141–80. PMID 10473346.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Created from PDB 1LMB
- ^ Myers L, Kornberg R. "Mediator of transcriptional regulation". Annu Rev Biochem. 69: 729–49. PMID 10966474.
- ^ Spiegelman B, Heinrich R (2004). "Biological control through regulated transcriptional coactivators". Cell. 119 (2): 157–67. PMID 15479634.
- ^ Li Z, Van Calcar S, Qu C, Cavenee W, Zhang M, Ren B (2003). "A global transcriptional regulatory role for c-Myc in Burkitt's lymphoma cells". Proc Natl Acad Sci U S A. 100 (14): 8164–9. PMID 12808131.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Pabo C, Sauer R. "Protein-DNA recognition". Annu Rev Biochem. 53: 293–321. PMID 6236744.
- ^ Created from PDB 1RVA
- ^ Bickle T, Krüger D (1993). "Biology of DNA restriction". Microbiol Rev. 57 (2): 434–50. PMID 8336674.
- ^ a b Doherty A, Suh S (2000). "Structural and mechanistic conservation in DNA ligases". Nucleic Acids Res. 28 (21): 4051–8. PMID 11058099.
- ^ Schoeffler A, Berger J (2005). "Recent advances in understanding structure-function relationships in the type II topoisomerase mechanism". Biochem Soc Trans. 33 (Pt 6): 1465–70. PMID 16246147.
- ^ Tuteja N, Tuteja R (2004). "Unraveling DNA helicases. Motif, structure, mechanism and function". Eur J Biochem. 271 (10): 1849–63. PMID 15128295.
- ^ a b Joyce C, Steitz T (1995). "Polymerase structures and function: variations on a theme?". J Bacteriol. 177 (22): 6321–9. PMID 7592405. Cite error: The named reference "Joyce" was defined multiple times with different content (see the help page).
- ^ Hubscher U, Maga G, Spadari S. "Eukaryotic DNA polymerases". Annu Rev Biochem. 71: 133–63. PMID 12045093.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Johnson A, O'Donnell M. "Cellular DNA replicases: components and dynamics at the replication fork". Annu Rev Biochem. 74: 283–315. PMID 15952889.
- ^ Tarrago-Litvak L, Andréola M, Nevinsky G, Sarih-Cottin L, Litvak S (1994). "The reverse transcriptase of HIV-1: from enzymology to therapeutic intervention". FASEB J. 8 (8): 497–503. PMID 7514143.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Martinez E (2002). "Multi-protein complexes in eukaryotic gene transcription". Plant Mol Biol. 50 (6): 925–47. PMID 12516863.
- ^ Orgel L. "Prebiotic chemistry and the origin of the RNA world" (PDF). Crit Rev Biochem Mol Biol. 39 (2): 99–123. PMID 15217990.
- ^ Davenport R (2001). "Ribozymes. Making copies in the RNA world". Science. 292 (5520): 1278. PMID 11360970.
- ^ Szathmáry E (1992). "What is the optimum size for the genetic alphabet?" (PDF). Proc Natl Acad Sci U S A. 89 (7): 2614–8. PMID 1372984.
- ^ Lindahl T (1993). "Instability and decay of the primary structure of DNA". Nature. 362 (6422): 709–15. PMID 8469282.
- ^ Vreeland R, Rosenzweig W, Powers D (2000). "Isolation of a 250 million-year-old halotolerant bacterium from a primary salt crystal". Nature. 407 (6806): 897–900. PMID 11057666.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Hebsgaard M, Phillips M, Willerslev E (2005). "Geologically ancient DNA: fact or artefact?". Trends Microbiol. 13 (5): 212–20. PMID 15866038.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Nickle D, Learn G, Rain M, Mullins J, Mittler J (2002). "Curiously modern DNA for a "250 million-year-old" bacterium". J Mol Evol. 54 (1): 134–7. PMID 11734907.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Dahm R (2005). "Friedrich Miescher and the discovery of DNA". Dev Biol. 278 (2): 274–88. PMID 15680349.
- ^ Levene P, (1919). "The structure of yeast nucleic acid". J Biol Chem. 40 (2): 415–24.
{{cite journal}}: CS1 maint: extra punctuation (link) - ^ Astbury W, (1947). "Nucleic acid". Symp. SOC. Exp. Bbl. 1 (66).
{{cite journal}}: CS1 maint: extra punctuation (link) - ^ Avery O, MacLeod C, McCarty M (1979). "Studies on the chemical nature of the substance inducing transformation of pneumococcal types. Inductions of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus type III". J Exp Med. 149 (2): 297–326. PMID 33226.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Hershey A, Chase M (1952). "Independent functions of viral protein and nucleic acid in growth of bacteriophage" (PDF). J Gen Physiol. 36 (1): 39–56. PMID 12981234.
- ^ a b Watson J.D. and Crick F.H.C. "A Structure for Deoxyribose Nucleic Acid". (PDF) Nature 171, 737 – 738 (1953). Accessed 13 Feb 2007.
- ^ Nature Archives Double Helix of DNA: 50 Years
- ^ Molecular Configuration in Sodium Thymonucleate. Franklin R. and Gosling R.G.Nature 171, 740 – 741 (1953)Nature Archives Full Text (PDF)
- ^ Molecular Structure of Deoxypentose Nucleic Acids. Wilkins M.H.F., A.R. Stokes A.R. & Wilson, H.R. Nature 171, 738 – 740 (1953)Nature Archives (PDF)
- ^ Evidence for 2-Chain Helix in Crystalline Structure of Sodium Deoxyribonucleate. Franklin R. and Gosling R.G. Nature 172, 156 – 157 (1953)Nature Archives,full text (PDF)
- ^ The Nobel Prize in Physiology or Medicine 1962 Nobelprize .org Accessed 22 Dec 06
- ^ Crick, F.H.C. On degenerate templates and the adaptor hypothesis (PDF). genome.wellcome.ac.uk (Lecture, 1955). Accessed 22 Dec 2006
- ^ Meselson M, Stahl F (1958). "The replication of DNA in Escherichia coli". Proc Natl Acad Sci U S A. 44 (7): 671–82. PMID 16590258.
- ^ The Nobel Prize in Physiology or Medicine 1968 Nobelprize.org Accessed 22 Dec 06
- ^ Houdebine L. "Transgenic animal models in biomedical research". Methods Mol Biol. 360: 163–202. PMID 17172731.
- ^ Daniell H, Dhingra A (2002). "Multigene engineering: dawn of an exciting new era in biotechnology". Curr Opin Biotechnol. 13 (2): 136–41. PMID 11950565.
- ^ Job D (2002). "Plant biotechnology in agriculture". Biochimie. 84 (11): 1105–10. PMID 12595138.
- ^ Collins A, Morton N (1994). "Likelihood ratios for DNA identification" (PDF). Proc Natl Acad Sci U S A. 91 (13): 6007–11. PMID 8016106.
- ^ Weir B, Triggs C, Starling L, Stowell L, Walsh K, Buckleton J (1997). "Interpreting DNA mixtures". J Forensic Sci. 42 (2): 213–22. PMID 9068179.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Jeffreys A, Wilson V, Thein S. "Individual-specific 'fingerprints' of human DNA". Nature. 316 (6023): 76–9. PMID 2989708.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Colin Pitchfork — first murder conviction on DNA evidence also clears the prime suspect Forensic Science Service Accessed 23 Dec 2006
- ^ "DNA Identification in Mass Fatality Incidents". National Institute of Justice. September 2006.
- ^ Baldi, Pierre. Brunak, Soren. Bioinformatics: The Machine Learning Approach MIT Press (2001) ISBN 978-0-262-02506-5
- ^ Gusfield, Dan. Algorithms on Strings, Trees, and Sequences: Computer Science and Computational Biology. Cambridge University Press, 15 January 1997. ISBN 978-0-521-58519-4.
- ^ Sjölander K (2004). "Phylogenomic inference of protein molecular function: advances and challenges". Bioinformatics. 20 (2): 170–9. PMID 14734307.
- ^ Mount DM (2004). Bioinformatics: Sequence and Genome Analysis (2 ed.). Cold Spring Harbor Laboratory Press. ISBN 0879697121.
{{cite book}}: Text "Cold Spring Harbor, NY" ignored (help); Text "location" ignored (help) - ^ Adleman L (1994). "Molecular computation of solutions to combinatorial problems". Science. 266 (5187): 1021–4. PMID 7973651.
- ^ Parker J (2003). "Computing with DNA". EMBO Rep. 4 (1): 7–10. PMID 12524509.
- ^ Ashish Gehani, Thomas LaBean and John Reif. DNA-Based Cryptography. Proceedings of the 5th DIMACS Workshop on DNA Based Computers, Cambridge, MA, USA, 14 – 15 June 1999.
- ^ Wray G (2002). "Dating branches on the tree of life using DNA". Genome Biol. 3 (1): REVIEWS0001. PMID 11806830.
- ^ Lost Tribes of Israel, NOVA, PBS airdate: 22 February 2000. Transcript available from PBS.org, (last accessed on 4 March 2006)
- ^ Kleiman, Yaakov. "The Cohanim/DNA Connection: The fascinating story of how DNA studies confirm an ancient biblical tradition". aish.com (January 13, 2000). Accessed 4 March 2006.
- ^ Bhattacharya, Shaoni. "Killer convicted thanks to relative's DNA". newscientist.com (20 April 2004). Accessed 22 Dec 06
Further reading[edit]
- Clayton, Julie. (Ed.). 50 Years of DNA, Palgrave MacMillan Press, 2003. ISBN 978-1-40-391479-8
- Judson, Horace Freeland. The Eighth Day of Creation: Makers of the Revolution in Biology, Cold Spring Harbor Laboratory Press, 1996. ISBN 978-0-87-969478-4
- Olby, Robert. The Path to The Double Helix: Discovery of DNA, first published in October 1974 by MacMillan, with foreword by Francis Crick; ISBN 978-0-48-668117-7; the definitive DNA textbook, revised in 1994, with a 9 page postscript.
- Ridley, Matt. Francis Crick: Discoverer of the Genetic Code (Eminent Lives) HarperCollins Publishers; 192 pp, ISBN 978-0-06-082333-7 2006
- Rose, Steven. The Chemistry of Life, Penguin, ISBN 978-0-14-027273-4.
- Watson, James D. and Francis H.C. Crick. A structure for Deoxyribose Nucleic Acid (PDF). Nature 171, 737 – 738, 25 April 1953.
- Watson, James D. DNA: The Secret of Life ISBN 978-0-375-41546-3.
- Watson, James D. The Double Helix: A Personal Account of the Discovery of the Structure of DNA (Norton Critical Editions). ISBN 978-0-393-95075-5
- Watson, James D. "Avoid boring people and other lessons from a life in science" New York: Random House. ISBN 978-0-375-421844 Parameter error in {{ISBN}}: checksum (0-375-41284-0)366pp 2007
- Calladine, Chris R.; Drew, Horace R.; Luisi, Ben F. and Travers, Andrew A. Understanding DNA, Elsevier Academic Press, 2003. ISBN 978-0-12155089-9
DVD[edit]
- DNA — The Story of the Pioneers who Changed the World, Windfall Films Production for Channel Four Television & PBS Thirteen-WNET — 2003, PAL [1], NTSC PBS Shop
- DNA interactive PAL [2], NTSC [3], [4]
- DNA: The Secret of Life Carolina Biological
- DNA — Secret of Photo 51 Rosalind Franklin — NOVA documentary (NTSC — Region 1?)
- Cracking the Code of Life NOVA documentary (NTSC — All Regions)
External links[edit]
{{Spoken Wikipedia|dna.ogg|2007-02-12}}
- [5] Crick's personal papers at Mandeville Special Collections Library, Geisel Library, University of California, San Diego
- DNA Interactive (requires Adobe Flash)
- DNA from the beginning
- Double Helix 1953 – 2003 National Centre for Biotechnology Education
- Double helix: 50 years of DNA, Nature
- Rosalind Franklin's contributions to the study of DNA
- U.S. National DNA Day — watch videos and participate in real-time chat with top scientists
- Genetic Education Modules for Teachers — DNA from the Beginning Study Guide
- Listen to Francis Crick and James Watson talking on the BBC in 1962, 1972, and 1974
- PDB Molecule of the Month DNA/Test
- DNA under electron microscope
- DNA at Curlie
- DNA Articles — articles and information collected from various sources
- DNA coiling to form chromosomes
- DISPLAR: DNA binding site prediction on protein
- Dolan DNA Learning Center
- Olby, R. (2003) "Quiet debut for the double helix" Nature 421 (January 23): 402 – 405.
- Basic animated guide to DNA cloning
ar:حمض نووي ريبي منقوص الأكسجين
az:Dezoksiribonuklein turşusu
zh-min-nan:DNA
bs:Dezoksiribonukleinska kiselina
bg:ДНК
ca:Àcid desoxiribonucleic
cs:DNA
cy:DNA
da:Dna
pdc:DNA
de:Desoxyribonukleinsäure
et:Desoksüribonukleiinhape
el:DNA
es:ADN
eo:DNA
eu:ADN
fa:دیانای
fr:Acide désoxyribonucléique
ga:ADN
gl:ADN
ko:DNA
hr:Deoksiribonukleinska kiselina
id:Asam deoksiribonukleat
it:DNA
he:DNA
ka:დეზოქსირიბონუკლეინის მჟავა
ht:ADN
la:Acidum deoxyribonucleinicum
lv:Dezoksiribonukleīnskābe
lt:Deoksiribonukleorūgštis
hu:DNS (biológia)
mk:ДНК
ms:DNA
nl:DNA
ja:デオキシリボ核酸
no:DNA
nn:Deoksyribonukleinsyre
pam:DNA
pl:Kwas deoksyrybonukleinowy
pt:DNA
ro:ADN
ru:Дезоксирибонуклеиновая кислота
sq:ADN
simple:DNA
sk:Deoxyribonukleová kyselina
sl:Deoksiribonukleinska kislina
sr:ДНК
sh:DNK
su:DNA
fi:DNA
sv:DNA
tl:DNA
ta:ஆக்சிஜனற்ற ரைபோ கரு அமிலம்
th:ดีเอ็นเอ
vi:DNA
tr:DNA
uk:ДНК
ur:ڈی این اے
yi:די ען עי
zh:脱氧核糖核酸

