User:Slj758/sandbox
Production of the Primary Transcript[edit]
The steps contributing to the production of the primary transcript involve a series of interactions that initiate the transcription of DNA within a cell's nucleus. Based on the needs of a given cell, certain DNA sequences are transcribed to produce a variety of RNA products. To initiate the transcription process in a cell's nucleus, DNA double helices are unwound and hydrogen bonds connecting compatible nucleic acids of DNA are broken to produce two unconnected single strands.[1] The DNA template to be used for transcription of the single-stranded primary transcript mRNA is then bound by an RNA polymerase at the promoter region.[2]
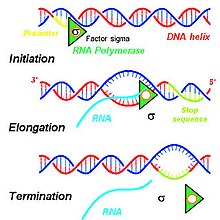
In eukaryotes, three kinds of RNA—rRNA, tRNA, and mRNA—are produced based on the activity of three distinct RNA polymerases, whereas, in prokaryotes, only one RNA polymerase exists to create all kinds of RNA molecules.[3] RNA polymerase II of eukaryotes transcribes the primary transcript mRNA from the antisense DNA template in the 5' to 3' direction, and the newly synthesized mRNA is complimentary to this template strand of DNA (see Figure).[1] RNA polymerase II constructs the primary transcript using a set of four specific ribonucleoside monophosphate residues (AMP, CMP, GMP, and UMP) that are added continuously to the 3' hydroxyl group on the 3' end of the growing mRNA.[1]
Studies of primary transcripts produced by RNA polymerase II reveal that a given primary transcript averages 7000 nucleotides in length, some growing as long as 20,000 nucleotides in length.[4] Inclusion of the exon and intron components within primary transcript sequences explain for the extreme size disparity between larger primary transcripts and smaller, modified mRNA ready for translation into protein.
Regulation of Primary Transcript Production[edit]
A number of factors contribute to the activation and inhibition of transcription and therefore regulate the production of primary transcript mRNA from a given DNA template. Activation of RNA polymerase activity to produce primary transcripts is often controlled by other DNA sequence elements called enhancers. The transcription factors that bind to enhancers recruit enzymes that alter nucleosome components to be either more or less accessible to RNA polymerase. The unique combinations of transcription factors that bind to enhancer DNA regions determine whether or not the gene that enhancer interacts with becomes activated or not.[5] The activation of transcription by RNA polymerase depends on whether or not the transcription elongation complex, itself consisting of a variety of transcription factors, can induce RNA polymerase to dissociate from the Mediator complex connecting an enhancer region to the promoter.[5]

Inhibition of RNA polymerase activity can also be regulated by DNA elements called silencers. Like enhancers, silencers may be located at locations farther up or downstream from the genes they regulate. These DNA sequences bind to factors that contribute to the destabilization of the initiation complex required to activate RNA polymerase, and therefore inhibit transcription.[6]
Histone modification by transcription factors is another key regulatory factor for transcription by RNA polymerase. Factors that lead to histone acetylation activate transcription while factors that lead to histone deacetylation inhibit transcription.[7] Acetylation of histones induces repulsion between negative components within nucleosomes, opening up space for RNA polymerase access. Deacetylation of histones favors more tightly coiled nucleosomes, inhibiting RNA polymerase access. In addition to acetylation patterns of histones, methylation patterns at promoter regions of DNA can regulate RNA polymerase access to a desired template. RNA polymerase is often incapable of synthesizing a primary transcript if the targeted gene's promoter region contains specific methylated cytosines— residues that hinder binding of transcription-activating factors and recruit other enzymes to stabilize a tightly bound nucleosome structure, excluding access to RNA polymerase.[5]
References[edit]
- ^ a b c T. Strachan; Andrew P. Read (January 2004). Human Molecular Genetics 3. Garland Science. pp. 16–17. ISBN 978-0-8153-4184-0. Cite error: The named reference "StrachanRead2004" was defined multiple times with different content (see the help page).
- ^ Alberts, B. "Molecular Biology of the Cell 3rd Edition". NCBI. New York: Garland Science.
- ^ Griffiths, AJF. "An Introduction to Genetic Analysis". NCBI. New York: W.H. Freeman.
- ^ Alberts, B. "Molecular Biology of the Cell 3rd Edition". NCBI. New York: Garland Science.
- ^ a b c Scott F. Gilbert (15 July 2013). Developmental Biology. Sinauer Associates, Incorporated. p. 38. ISBN 978-1-60535-173-5. Cite error: The named reference "Gilbert2013" was defined multiple times with different content (see the help page).
- ^ Brown, T.A. "Genomes 2nd Edition". NCBI. Oxford: Wiley-Liss.
- ^ Lodish, H. "Molecular Cell Biology". NCBI. New York: W.H. Freeman.

