Bilirubin oxidase
| bilirubin oxidase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
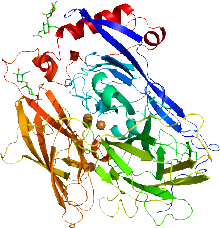 Cartoon representation of the X-ray structure of bilirubin oxidase from Myrothecium verrucaria based on PDB accession code 2xll. The protein ribbon is rainbow colored with the N-terminus in blue and the C-terminus in red. The four copper atoms are shown as spheres and the glycans shown as sticks. | |||||||||
| Identifiers | |||||||||
| EC no. | 1.3.3.5 | ||||||||
| CAS no. | 80619-01-8 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
In enzymology, a bilirubin oxidase, BOD or BOx, (EC 1.3.3.5) is an enzyme encoded by a gene in various organisms that catalyzes the chemical reaction
- 2 bilirubin + O2 2 biliverdin + 2 H2O
This enzyme belongs to the family of oxidoreductases, to be specific those acting on the CH-CH group of donor with oxygen as acceptor. The systematic name of this enzyme class is bilirubin:oxygen oxidoreductase. This enzyme is also called bilirubin oxidase M-1. This enzyme participates in porphyrin and chlorophyll metabolism. It is widely studied as a catalyst for oxygen reduction.[1]
Two structures of bilirubin oxidase from the ascomycete Myrothecium verrucaria have been deposited in the Protein Data Bank (accession codes 3abg and 2xll).[2][3]
The active site consists of four copper centers, reminiscent of laccase. These centers are classified as type I (cys, met, his, his), type II (3his), and two type III (2his).[4] The latter two centers are arranged in a trinuclear copper cluster forming the active site for oxygen reduction.[5] The type I copper center is the primary electron acceptor and the site for the reduction of bilirubin.
References
[edit]- ^ Mano N, Edembe L (December 2013). "Bilirubin oxidases in bioelectrochemistry: features and recent findings". Biosensors & Bioelectronics. 50: 478–485. doi:10.1016/j.bios.2013.07.014. PMID 23911663.
- ^ Mizutani K, Toyoda M, Sagara K, Takahashi N, Sato A, Kamitaka Y, et al. (July 2010). "X-ray analysis of bilirubin oxidase from Myrothecium verrucaria at 2.3 A resolution using a twinned crystal". Acta Crystallographica. Section F, Structural Biology and Crystallization Communications. 66 (Pt 7): 765–770. doi:10.1107/S1744309110018828. PMC 2898457. PMID 20606269.
- ^ Cracknell JA, McNamara TP, Lowe ED, Blanford CF (July 2011). "Bilirubin oxidase from Myrothecium verrucaria: X-ray determination of the complete crystal structure and a rational surface modification for enhanced electrocatalytic O2 reduction". Dalton Transactions. 40 (25): 6668–6675. doi:10.1039/c0dt01403f. PMID 21544308.
- ^ Jones SM, Solomon EI (March 2015). "Electron transfer and reaction mechanism of laccases". Cellular and Molecular Life Sciences. 72 (5): 869–883. doi:10.1007/s00018-014-1826-6. PMC 4323859. PMID 25572295.
- ^ Mano N, de Poulpiquet A (March 2018). "O2 Reduction in Enzymatic Biofuel Cells" (PDF). Chemical Reviews. 118 (5): 2392–2468. doi:10.1021/acs.chemrev.7b00220. PMID 28930449.
Further reading
[edit]- Murao S, Tanaka N (1981). "A new enzyme bilirubin oxidase produced by Myrothecium verrucaria MT-1". Agricultural and Biological Chemistry. 45 (10): 2383–2384. doi:10.1271/bbb1961.45.2383.
- Tanaka N, Murao S (1985). "Reaction of bilirubin oxidase produced by Myrothecium verrucaria MT-1". Agricultural and Biological Chemistry. 49 (3): 843–844. doi:10.1271/bbb1961.49.843.

