Ciliopathy
| Ciliopathy | |
|---|---|
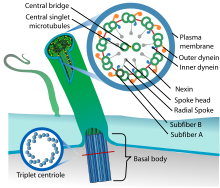 | |
| Eukaryotic cilium | |
| Specialty | Medical genetics |
A ciliopathy is any genetic disorder that affects the cellular cilia or the cilia anchoring structures, the basal bodies,[1] or ciliary function.[2] Primary cilia are important in guiding the process of development, so abnormal ciliary function while an embryo is developing can lead to a set of malformations that can occur regardless of the particular genetic problem.[3] The similarity of the clinical features of these developmental disorders means that they form a recognizable cluster of syndromes, loosely attributed to abnormal ciliary function and hence called ciliopathies. Regardless of the actual genetic cause, it is clustering of a set of characteristic physiological features which define whether a syndrome is a ciliopathy.
Although ciliopathies are usually considered to involve proteins that localize to motile and/or immotile (primary) cilia or centrosomes, it is possible for ciliopathies to be associated with unexpected proteins such as XPNPEP3, which localizes to mitochondria but is believed to affect ciliary function through proteolytic cleavage of ciliary proteins.[4]
Significant advances in understanding the importance of cilia were made in the mid-1990s. For example, the discovery of the role of cilia in embryonic development, identification of ciliary defects in genetic disorders such as Polycystic kidney disease, Bardet–Biedl syndrome and Primary ciliary dyskinesia.[5][6] However, the physiological role that this organelle plays in most tissues remains elusive. Additional studies of how ciliary dysfunction can lead to such severe disease and developmental pathologies is still a subject of current research.[7]
Signs and symptoms
[edit]A wide variety of symptoms are potential clinical features of ciliopathy. The signs most exclusive to a ciliopathy, in descending order of exclusivity, are:[8]: 138
- Dandy–Walker malformation (cerebellar vermis hypoplasia, usually with hydrocephalus)
- Agenesis of the corpus callosum
- Situs inversus
- Posterior encephalocele
- Polycystic kidneys
- Postaxial polydactyly
- Liver disease
- Retinitis pigmentosa
- Intellectual disability
A case with polycystic ovary syndrome, multiple subcutaneous cysts, renal function impairment, Caroli disease and liver cirrhosis due to ciliopathy has been described.[9]
Phenotypes sometimes associated with ciliopathies can include:[8]
- Anencephaly
- Breathing abnormalities
- Cerebellar vermis hypoplasia
- Diabetes
- Exencephaly
- Eye movement abnormalities
- Hydrocephalus
- Hypoplasia of the corpus callosum
- Hypotonia
- Infertility
- Cognitive impairment/defects
- Obesity[10]
- Other polydactyly
- Respiratory dysfunction
- Renal cystic disease
- Retinal degeneration
- Sensorineural deafness
- Spina bifida
Pathophysiology
[edit]"In effect, the motile cilium is a nanomachine composed of perhaps over 600 proteins in molecular complexes, many of which also function independently as nanomachines." Cilia "function as mechano- or chemosensors and as a cellular global positioning system to detect changes in the surrounding environment." For example, ciliary signaling plays a role in the initiation of cellular replacement after cell damage.[11]
In addition to this sensory role mediating specific signaling cues, cilia play "a secretory role in which a soluble protein is released to have an effect downstream of the fluid flow" in epithelial cells, and can of course mediate fluid flow directly in the case of motile cilia.[1] Primary cilia in the retina play a role in transferring nourishment to the non-vascularized rod and cone cells from the pigment epithelial vascularized cells several micrometres behind the surface of the retina.
Signal transduction pathways involved include the Hedgehog signaling pathway and the Wnt signaling pathway.[12]
Dysfunctional cilia can lead to:
- Chemosensation abnormalities,[13] typically via ciliated epithelial cellular dysfunction.[1]
- Defective thermosensation or mechanosensation,[14] often via ciliated epithelial cellular dysfunction.[1]
- Cellular motility dysfunction[13]
- Issues with displacement of extracellular fluid[13]
- Paracrine signal transduction abnormalities[1][13]
In organisms of normal health, cilia are critical for:[15]
Genetics
[edit]"Just as different genes can contribute to similar diseases, so the same genes and families of genes can play a part in a range of different diseases." For example, in just two of the diseases caused by malfunctioning cilia, Meckel–Gruber syndrome and Bardet–Biedl syndrome, patients who carry mutations in genes associated with both diseases "have unique symptoms that are not seen in either condition alone." The genes linked to the two different conditions "interact with each other during development." Systems biologists are endeavoring to define functional modules containing multiple genes and then look at disorders whose phenotypes fit into such modules.[16]
A particular phenotype can overlap "considerably with several conditions (ciliopathies) in which primary cilia are also implicated in pathogenicity. One emerging aspect is the wide spectrum of ciliopathy gene mutations found within different diseases."[10]
Additionally, clinical presentations of patients with identical mutation can differ, suggesting the role of genetic modifiers.[17]
As of 2017, 187 ciliopathy associated genes have been confirmed, while the roles of further 241 candidate genes are still being investigated.[18]
A common way to identify ciliopathies such as ADPKD and ARPKD which have known genetic causes, is through linkage analysis direct mutation screening.[19] Other techniques, such as gene panels and whole-exome sequencing and whole genome sequencing can also be used to provide distinct advantages.[19][20] Gene panels analyse specific sets of genes and can be more comprehensive than single gene or direct mutation screening. Whole-exome/genome sequencing can screen for heterozygous carriers, and detect novel/rare variations.[19][21]
Mutations in the PKD1 and PKD2 genes which encode for polycystin-1 and polycistin-2 respectively are known to be causes of ADPKD, a ciliopathy that presents with the formation and growth of cysts in the kidneys, leading to renal failure.[22]
List of ciliopathies
[edit]"The phenotypic parameters that define a ciliopathy may be used to both recognize the cellular basis of a number of genetic disorders and to facilitate the diagnosis and treatment of some diseases of unknown" cause.[8]
Known ciliopathies
[edit]Likely ciliopathies
[edit]Possible ciliopathies
[edit]History
[edit]In 1674–1677, the Dutch scientist Antonie van Leeuwenhoek changed humanity's perspective on the world with his discovery of "animalcules" in rainwater, along with their tiny appendages known as cilia today. It was marked as the first recorded observation of single-celled organisms and their locomotive structures.[30]
In the late 19th century, Karl Ernst von Baer's groundbreaking work in embryonic development laid the foundation for modern developmental biology.[31] Through meticulous observations, von Baer provided invaluable insights into tissue and organ formation during development, including the early stages of embryogenesis and the development of cilia-bearing tissues.[32] While von Baer may not have fully appreciated the significance of cilia at the time, his observations likely included their presence in embryonic tissues. Cilia - crucial for cell signaling, tissue development, and left-right asymmetry, are now recognized as ancient organelles with essential roles in development.[33] Von Baer's concept of embryonic recapitulation, despite refinement, underscores the evolutionary conservation of developmental processes, including ciliary function. Today, von Baer's legacy inspires ongoing research into embryology and developmental biology, particularly in understanding ciliary biology and its relevance to ciliopathies, where defects in ciliary structure or function lead to developmental disorder.[34]
Although non-motile or primary cilia were first described in 1898, they were largely ignored by biologists. However, microscopists continued to document their presence in the cells of most vertebrate organisms. The primary cilium was long considered—with few exceptions—to be a largely useless evolutionary vestige, a vestigial organelle. Recent research has revealed that cilia are essential to many of the body's organs.[35] These primary cilia play important roles in chemosensation, mechanosensation, and thermosensation. Cilia may thus be "viewed as sensory cellular antennae that coordinate a large number of cellular signaling pathways, sometimes coupling the signaling to ciliary motility or alternatively to cell division and differentiation."[11]
Recent advances in mammalian genetic research have made possible the understanding of a molecular basis for a number of dysfunctional mechanisms in both motile and primary cilia structures of the cell.[36] A number of critical developmental signaling pathways essential to cellular development have been discovered. These are principally but not exclusively found in the non-motile or primary cilia. A number of common observable characteristics of mammalian genetic disorders and diseases are caused by ciliary dysgenesis and dysfunction. Once identified, these characteristics thus describe a set of hallmarks of a ciliopathy.[8]
Cilia have recently been implicated in a wide variety of human genetic diseases by "the discovery that numerous proteins involved in mammalian disease localize to the basal bodies and cilia." For example, in just a single area of human disease physiology, cystic renal disease, cilia-related genes and proteins have been identified to have causal effect in polycystic kidney disease, nephronophthisis, Senior–Løken syndrome type 5, orofaciodigital syndrome type 1 and Bardet–Biedl syndrome.[7]
References
[edit]- ^ a b c d e f g Adams M, Smith UM, Logan CV, Johnson CA (2008). "Recent advances in the molecular pathology, cell biology and genetics of ciliopathies". Journal of Medical Genetics. 45 (5): 257–267. doi:10.1136/jmg.2007.054999. PMID 18178628.
- ^ Lee JH, Gleeson JG (May 2010). "The role of primary cilia in neuronal function". Neurobiol. Dis. 38 (2): 167–72. doi:10.1016/j.nbd.2009.12.022. PMC 2953617. PMID 20097287.
- ^ Powles-Glover N (September 2014). "Cilia and ciliopathies: classic examples linking phenotype and genotype-an overview". Reproductive Toxicology (Elmsford, N.Y.). 48: 98–105. Bibcode:2014RepTx..48...98P. doi:10.1016/j.reprotox.2014.05.005. PMID 24859270.
- ^ Hurd TW, Hildebrandt F (2011). "Mechanisms of Nephronophthisis and Related Ciliopathies". Nephron Exp. Nephrol. 118 (1): e9–e14. doi:10.1159/000320888. PMC 2992643. PMID 21071979.
- ^ Cowley BD, Bissler JJ, eds. (2018). Polycystic Kidney Disease. New York: Springer. p. 87. doi:10.1007/978-1-4939-7784-0. ISBN 978-1-4939-7782-6.
- ^ Kenny TD, Beales PL (2014). Ciliopathies: a reference for clinicians. Oxford: Oxford University Press. ISBN 978-0-19-965876-3.
- ^ a b c d e f g Davenport JR (2005). "An incredible decade for the primary cilium: A look at a once-forgotten organelle". AJP: Renal Physiology. 289 (6): F1159–F1169. doi:10.1152/ajprenal.00118.2005. PMID 16275743.
- ^ a b c d e f g h i j k l m n o Badano JL, Mitsuma N, Beales PL, Katsanis N (2006). "The ciliopathies: an emerging class of human genetic disorders". Annu Rev Genom Hum Genet. 7: 125–48. doi:10.1146/annurev.genom.7.080505.115610. PMID 16722803.
- ^ Tan K, Liu P, Pang L, Yang W, Hou F (2018) A human ciliopathy with polycystic ovarian syndrome and multiple subcutaneous cysts: A rare case report. Medicine (Baltimore) 97(50)
- ^ a b c d e f Ross A, PL Beales, J Hill (2008). "The Clinical, Molecular, and Functional Genetics of Bardet-Biedl Syndrome". Genetics of Obesity Syndromes. Oxford University Press. p. 177. ISBN 978-0-19-530016-1. Retrieved 1 July 2009.
- ^ a b Satir P, Søren T. Christensen (26 March 2008). "Structure and function of mammalian cilia". Histochemistry and Cell Biology. 129 (6). Springer Berlin / Heidelberg: 687–693. doi:10.1007/s00418-008-0416-9. PMC 2386530. PMID 18365235. 1432-119X.
- ^ D'Angelo A, Franco B (2009). "The dynamic cilium in human diseases". Pathogenetics. 2 (1): 3. doi:10.1186/1755-8417-2-3. PMC 2694804. PMID 19439065.
- ^ a b c d "Ciliary proteome database, v3". Database introduction. Johns Hopkins University. 2008. Archived from the original on 29 April 2019. Retrieved 7 January 2009.
- ^ Tan PL, Barr T, Inglis PN, et al. (2007). "Loss of Bardet–Biedl syndrome proteins causes defects in peripheral sensory innervation and function". Proc. Natl. Acad. Sci. U.S.A. 104 (44): 17524–9. doi:10.1073/pnas.0706618104. PMC 2077289. PMID 17959775.
- ^ of organs The Ciliary Proteome Archived 29 April 2019 at the Wayback Machine, Ciliaproteome V3.0 – Home Page, accessed 11 June 2010.
- ^ Hayden EC (2008). "Biological tools revamp disease classification". Nature. 453 (7196): 709. doi:10.1038/453709a. PMID 18528360.
- ^ Walia S, Fishman GA, Swaroop A, Branham KE, Lindeman M, Othman M, et al. (March 2008). "Discordant phenotypes in fraternal twins having an identical mutation in exon ORF15 of the RPGR gene". Archives of Ophthalmology. 126 (3): 379–384. doi:10.1001/archophthalmol.2007.72. ISSN 0003-9950. PMID 18332319.
- ^ Reiter JF, Leroux MR (September 2017). "Genes and molecular pathways underpinning ciliopathies". Nature Reviews. Molecular Cell Biology. 18 (9): 533–547. doi:10.1038/nrm.2017.60. ISSN 1471-0072. PMC 5851292. PMID 28698599.
- ^ a b c Modarage K, Malik SA, Goggolidou P (10 January 2022). "Molecular Diagnostics of Ciliopathies and Insights Into Novel Developments in Diagnosing Rare Diseases". British Journal of Biomedical Science. 79: 10221. doi:10.3389/bjbs.2021.10221. ISSN 0967-4845. PMC 8915726. PMID 35996505.
- ^ "Genetic testing for". Blueprint Genetics. Retrieved 15 February 2024.
- ^ Shamseldin HE, Shaheen R, Ewida N, Bubshait DK, Alkuraya H, Almardawi E, et al. (June 2020). "The morbid genome of ciliopathies: an update". Genetics in Medicine. 22 (6): 1051–1060. doi:10.1038/s41436-020-0761-1. ISSN 1530-0366. PMID 32055034.
- ^ "PKD1 – an overview | ScienceDirect Topics". www.sciencedirect.com. Retrieved 15 February 2024.
- ^ a b c d e f g h i j k l m n o p q r s t u v w x y z aa ab ac ad ae af ag ah ai aj ak al am an ao ap aq ar as at au av aw ax ay az ba bb bc bd be bf bg bh bi bj bk bl bm bn bo bp bq br bs bt bu bv bw bx by bz ca cb cc cd ce cf cg ch ci cj ck cl cm cn co Baker K, Beales PL (2009). "Making sense of cilia in disease: The human ciliopathies". American Journal of Medical Genetics Part C: Seminars in Medical Genetics. 151C (4): 281–295. doi:10.1002/ajmg.c.30231. ISSN 1552-4876. PMID 19876933.
- ^ Kyttälä M (May 2006). "Identification of the Meckel Syndrome Gene (MKS1) Exposes a Novel Ciliopathy" (PDF). National Public Health Institute. Archived from the original (PDF) on 21 July 2006. Retrieved 6 July 2008.
- ^ Gunay-Aygun M (November 2009). "Liver and Kidney Disease in Ciliopathies". Am J Med Genet C Semin Med Genet. 151C (4): 296–306. doi:10.1002/ajmg.c.30225. PMC 2919058. PMID 19876928.
- ^ Gene therapy rescues cilia defects and restores olfactory function in a mammalian ciliopathy model
- ^ Watnick T, Germino G (August 2003). "From cilia to cyst". Nat. Genet. 34 (4): 355–6. doi:10.1038/ng0803-355. PMID 12923538.
- ^ Delgado-Escueta AV (2007). "Advances in Genetics of Juvenile Myoclonic Epilepsies". Epilepsy Curr. 7 (3): 61–7. doi:10.1111/j.1535-7511.2007.00171.x. PMC 1874323. PMID 17520076.
- ^ Khanna H, Davis EE, Murga-Zamalloa CA, Estrada-Cuzcano A, Lopez I, Den Hollander AI, et al. (2009). "A common allele in RPGRIP1L is a modifier of retinal degeneration in ciliopathies". Nature Genetics. 41 (6): 739–745. doi:10.1038/ng.366. PMC 2783476. PMID 19430481.
- ^ Brown JM, Witman GB (1 December 2014). "Cilia and Diseases". BioScience. 64 (12): 1126–1137. doi:10.1093/biosci/biu174. ISSN 1525-3244. PMC 4420261. PMID 25960570.
- ^ Brauckmann S (2012). "Karl Ernst von Baer (1792–1876) and Evolution". The International Journal of Developmental Biology. 56 (9): 653–660. doi:10.1387/ijdb.120018sb. ISSN 0214-6282. PMID 23319342.
- ^ Abzhanov A (2013). "von Baer's law for the ages: lost and found principles of developmental evolution". Trends in Genetics. 29 (12): 712–722. doi:10.1016/j.tig.2013.09.004. ISSN 0168-9525. PMID 24120296. S2CID 9158143.
- ^ Sreekumar V, Norris DP (1 June 2019). "Cilia and development". Current Opinion in Genetics & Development. Molecular and genetic basis of disease. 56: 15–21. doi:10.1016/j.gde.2019.05.002. ISSN 0959-437X. PMID 31201996. S2CID 189898579.
- ^ Bisgrove BW, Yost HJ (1 November 2006). "The roles of cilia in developmental disorders and disease". Development. 133 (21): 4131–4143. doi:10.1242/dev.02595. ISSN 1477-9129. PMID 17021045. S2CID 9975220.
- ^ Gardiner MB (September 2005). "The Importance of Being Cilia". HHMI Bulletin. 18 (2). Howard Hughes Medical Institute. Archived from the original on 11 March 2010. Retrieved 26 July 2008.
- ^ Lancaster MA, Gleeson JG (June 2009). "The primary cilium as a cellular signaling center: lessons from disease". Curr. Opin. Genet. Dev. 19 (3): 220–9. doi:10.1016/j.gde.2009.04.008. PMC 2953615. PMID 19477114.
External links
[edit]- The Ciliary Proteome Web Page at Johns Hopkins Archived 29 April 2019 at the Wayback Machine
