Serpin
| Serpin (serine protease inhibitor) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
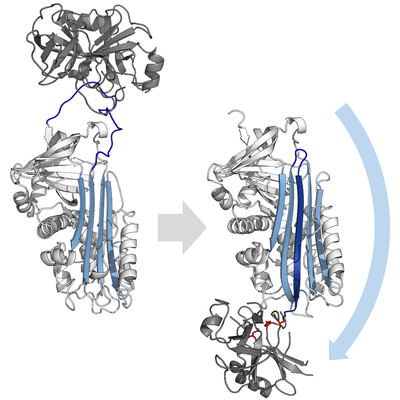
 A serpin (white) with its 'reactive centre loop' (blue) bound to a protease (grey). Once the protease attempts catalysis it will be irreversibly inhibited. (PDB: 1K9O) | |||||||||||
| Identifiers | |||||||||||
| Symbol | Serpin, SERPIN (root symbol of family) | ||||||||||
| Pfam | PF00079 | ||||||||||
| InterPro | IPR000215 | ||||||||||
| PROSITE | PDOC00256 | ||||||||||
| SCOP2 | 1hle / SCOPe / SUPFAM | ||||||||||
| CDD | cd00172 | ||||||||||
| |||||||||||
Serpins are a superfamily of proteins with similar structures that were first identified for their protease inhibition activity and are found in all kingdoms of life.[1][2] The acronym serpin was originally coined because the first serpins to be identified act on chymotrypsin-like serine proteases (serine protease inhibitors).[3][4][5] They are notable for their unusual mechanism of action, in which they irreversibly inhibit their target protease by undergoing a large conformational change to disrupt the target's active site.[6][7] This contrasts with the more common competitive mechanism for protease inhibitors that bind to and block access to the protease active site.[8][9]
Protease inhibition by serpins controls an array of biological processes, including coagulation and inflammation, and consequently these proteins are the target of medical research.[10] Their unique conformational change also makes them of interest to the structural biology and protein folding research communities.[7][8] The conformational-change mechanism confers certain advantages, but it also has drawbacks: serpins are vulnerable to mutations that can result in serpinopathies such as protein misfolding and the formation of inactive long-chain polymers.[11][12] Serpin polymerisation not only reduces the amount of active inhibitor, but also leads to accumulation of the polymers, causing cell death and organ failure.[10]
Although most serpins control proteolytic cascades, some proteins with a serpin structure are not enzyme inhibitors, but instead perform diverse functions such as storage (as in egg white—ovalbumin), transport as in hormone carriage proteins (thyroxine-binding globulin, cortisol-binding globulin) and molecular chaperoning (HSP47).[9] The term serpin is used to describe these members as well, despite their non-inhibitory function, since they are evolutionarily related.[1]
History
[edit]Protease inhibitory activity in blood plasma was first reported in the late 1800s,[13] but it was not until the 1950s that the serpins antithrombin and alpha 1-antitrypsin were isolated,[14] with the subsequent recognition of their close family homology in 1979.[15][16] That they belonged to a new protein family became apparent on their further alignment with the non-inhibitory egg-white protein ovalbumin, to give what was initially called the alpha1-antitrypsin-antithrombin III-ovalbumin superfamily of serine proteinase inhibitors,[17] but was subsequently succinctly renamed as the Serpins.[18] The initial characterisation of the new family centred on alpha1-antitrypsin, a serpin present in high concentration in blood plasma, the common genetic disorder of which was shown to cause a predisposition to the lung disease emphysema[19] and to liver cirrhosis.[20] The identification of the S and Z mutations[21][22] responsible for the genetic deficiency and the subsequent sequence alignments of alpha1-antitrypsin and antithrombin in 1982 led to the recognition of the close homologies of the active sites of the two proteins,[23][24] centred on a methionine[25] in alpha1-antitrypsin as an inhibitor of tissue elastase and on arginine in antithrombin[26] as an inhibitor of thrombin.[27]
The critical role of the active centre residue in determining the specificity of inhibition of serpins was unequivocally confirmed by the finding that a natural mutation of the active centre methionine in alpha1-antitrypsin to an arginine, as in antithrombin, resulted in a severe bleeding disorder.[28] This active-centre specificity of inhibition was also evident in the many other families of protease inhibitors[7] but the serpins differed from them in being much larger proteins and also in possessing what was soon apparent as an inherent ability to undergo a change in shape. The nature of this conformational change was revealed with the determination in 1984 of the first crystal structure of a serpin, that of post-cleavage alpha1-antitrypsin.[29] This together with the subsequent solving of the structure of native (uncleaved) ovalbumin[30] indicated that the inhibitory mechanism of the serpins involved a remarkable conformational shift, with the movement of the exposed peptide loop containing the reactive site and its incorporation as a middle strand in the main beta-pleated sheet that characterises the serpin molecule.[31][32] Early evidence of the essential role of this loop movement in the inhibitory mechanism came from the finding that even minor aberrations in the amino acid residues that form the hinge of the movement in antithrombin resulted in thrombotic disease.[31][33] Ultimate confirmation of the linked displacement of the target protease by this loop movement was provided in 2000 by the structure of the post-inhibitory complex of alpha1-antitrypsin with trypsin,[6] showing how the displacement results in the deformation and inactivation of the attached protease. Subsequent structural studies have revealed an additional advantage of the conformational mechanism[34] in allowing the subtle modulation of inhibitory activity, as notably seen at tissue level[35] with the functionally diverse serpins in human plasma.
Over 1000 serpins have now been identified, including 36 human proteins, as well as molecules in all kingdoms of life—animals, plants, fungi, bacteria, and archaea—and some viruses.[36][37][38] The central feature of all is a tightly conserved framework, which allows the precise alignment of their key structural and functional components based on the template structure of alpha1-antitrypsin.[39] In the 2000s, a systematic nomenclature was introduced in order to categorise members of the serpin superfamily based on their evolutionary relationships.[1] Serpins are therefore the largest and most diverse superfamily of protease inhibitors.[40]
Activity
[edit]
Most serpins are protease inhibitors, targeting extracellular, chymotrypsin-like serine proteases. These proteases possess a nucleophilic serine residue in a catalytic triad in their active site. Examples include thrombin, trypsin, and human neutrophil elastase.[41] Serpins act as irreversible, suicide inhibitors by trapping an intermediate of the protease's catalytic mechanism.[6]
Some serpins inhibit other protease classes, typically cysteine proteases, and are termed "cross-class inhibitors". These enzymes differ from serine proteases in that they use a nucleophilic cysteine residue, rather than a serine, in their active site.[42] Nonetheless, the enzymatic chemistry is similar, and the mechanism of inhibition by serpins is the same for both classes of protease.[43] Examples of cross-class inhibitory serpins include serpin B4 a squamous cell carcinoma antigen 1 (SCCA-1) and the avian serpin myeloid and erythroid nuclear termination stage-specific protein (MENT), which both inhibit papain-like cysteine proteases.[44][45][46]
Biological function and localization
[edit]Protease inhibition
[edit]Approximately two-thirds of human serpins perform extracellular roles, inhibiting proteases in the bloodstream in order to modulate their activities. For example, extracellular serpins regulate the proteolytic cascades central to blood clotting (antithrombin), the inflammatory and immune responses (antitrypsin, antichymotrypsin, and C1-inhibitor) and tissue remodelling (PAI-1).[9] By inhibiting signalling cascade proteases, they can also affect development.[47][48] The table of human serpins (below) provides examples of the range of functions performed by human serpin, as well as some of the diseases that result from serpin deficiency.
The protease targets of intracellular inhibitory serpins have been difficult to identify, since many of these molecules appear to perform overlapping roles. Further, many human serpins lack precise functional equivalents in model organisms such as the mouse. Nevertheless, an important function of intracellular serpins may be to protect against the inappropriate activity of proteases inside the cell.[49] For example, one of the best-characterised human intracellular serpins is Serpin B9, which inhibits the cytotoxic granule protease granzyme B. In doing so, Serpin B9 may protect against inadvertent release of granzyme B and premature or unwanted activation of cell death pathways.[50]
Some viruses use serpins to disrupt protease functions in their host. The cowpox viral serpin CrmA (cytokine response modifier A) is used in order to avoid inflammatory and apoptotic responses of infected host cells. CrmA increases infectivity by suppressing its host's inflammatory response through inhibition of IL-1 and IL-18 processing by the cysteine protease caspase-1.[51] In eukaryotes, a plant serpin inhibits both metacaspases[52] and a papain-like cysteine protease.[53]
Non-inhibitory roles
[edit]Non-inhibitory extracellular serpins also perform a wide array of important roles. Thyroxine-binding globulin and transcortin transport the hormones thyroxine and cortisol, respectively.[54][55] The non-inhibitory serpin ovalbumin is the most abundant protein in egg white. Its exact function is unknown, but it is thought to be a storage protein for the developing foetus.[56] Heat shock serpin 47 is a chaperone, essential for proper folding of collagen. It acts by stabilising collagen's triple helix whilst it is being processed in the endoplasmic reticulum.[57]
Some serpins are both protease inhibitors and perform additional roles. For example, the nuclear cysteine protease inhibitor MENT, in birds also acts as a chromatin remodelling molecule in a bird's red blood cells.[45][58]
Structure
[edit]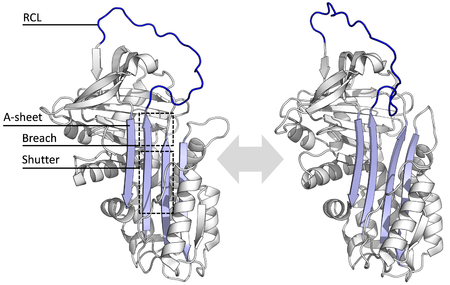
All serpins share a common structure (or fold), despite their varied functions. All typically have three β-sheets (named A, B and C) and eight or nine α-helices (named hA–hI).[29][30] The most significant regions to serpin function are the A-sheet and the reactive centre loop (RCL). The A-sheet includes two β-strands that are in a parallel orientation with a region between them called the 'shutter', and upper region called the 'breach'. The RCL forms the initial interaction with the target protease in inhibitory molecules. Structures have been solved showing the RCL either fully exposed or partially inserted into the A-sheet, and serpins are thought to be in dynamic equilibrium between these two states.[8] The RCL also only makes temporary interactions with the rest of the structure, and is therefore highly flexible and exposed to the solvent.[8]
The serpin structures that have been determined cover several different conformations, which has been necessary for the understanding of their multiple-step mechanism of action. Structural biology has therefore played a central role in the understanding of serpin function and biology.[8]
Conformational change and inhibitory mechanism
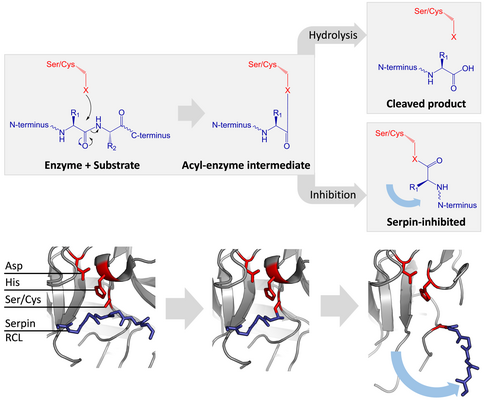
[edit]Inhibitory serpins do not inhibit their target proteases by the typical competitive (lock-and-key) mechanism used by most small protease inhibitors (e.g. Kunitz-type inhibitors). Instead, serpins use an unusual conformational change, which disrupts the structure of the protease and prevents it from completing catalysis. The conformational change involves the RCL moving to the opposite end of the protein and inserting into β-sheet A, forming an extra antiparallel β-strand. This converts the serpin from a stressed state, to a lower-energy relaxed state (S to R transition).[7][8][61]
Serine and cysteine proteases catalyse peptide bond cleavage by a two-step process. Initially, the catalytic residue of the active site triad performs a nucleophilic attack on the peptide bond of the substrate. This releases the new N-terminus and forms a covalent ester-bond between the enzyme and the substrate.[7] This covalent complex between enzyme and substrate is called an acyl-enzyme intermediate. For standard substrates, the ester bond is hydrolysed and the new C-terminus is released to complete catalysis. However, when a serpin is cleaved by a protease, it rapidly undergoes the S to R transition before the acyl-enzyme intermediate is hydrolysed.[7] The efficiency of inhibition depends on fact that the relative kinetic rate of the conformational change is several orders of magnitude faster than hydrolysis by the protease.
Since the RCL is still covalently attached to the protease via the ester bond, the S to R transition pulls protease from the top to the bottom of the serpin and distorts the catalytic triad. The distorted protease can only hydrolyse the acyl enzyme intermediate extremely slowly and so the protease remains covalently attached for days to weeks.[6] Serpins are classed as irreversible inhibitors and as suicide inhibitors since each serpin protein permanently inactivates a single protease, and can only function once.[7]
Allosteric activation
[edit]
The conformational mobility of serpins provides a key advantage over static lock-and-key protease inhibitors.[34] In particular, the function of inhibitory serpins can be regulated by allosteric interactions with specific cofactors. The X-ray crystal structures of antithrombin, heparin cofactor II, MENT and murine antichymotrypsin reveal that these serpins adopt a conformation wherein the first two amino acids of the RCL are inserted into the top of the A β-sheet. The partially inserted conformation is important because co-factors are able to conformationally switch certain partially inserted serpins into a fully expelled form.[62][63] This conformational rearrangement makes the serpin a more effective inhibitor.
The archetypal example of this situation is antithrombin, which circulates in plasma in a partially inserted relatively inactive state. The primary specificity determining residue (the P1 arginine) points toward the body of the serpin and is unavailable to the protease. Upon binding a high-affinity pentasaccharide sequence within long-chain heparin, antithrombin undergoes a conformational change, RCL expulsion, and exposure of the P1 arginine. The heparin pentasaccharide-bound form of antithrombin is, thus, a more effective inhibitor of thrombin and factor Xa.[64][65] Furthermore, both of these coagulation proteases also contain binding sites (called exosites) for heparin. Heparin, therefore, also acts as a template for binding of both protease and serpin, further dramatically accelerating the interaction between the two parties. After the initial interaction, the final serpin complex is formed and the heparin moiety is released. This interaction is physiologically important. For example, after injury to the blood vessel wall, heparin is exposed, and antithrombin is activated to control the clotting response. Understanding of the molecular basis of this interaction enabled the development of Fondaparinux, a synthetic form of Heparin pentasaccharide used as an anti-clotting drug.[66][67]
Latent conformation
[edit]
Certain serpins spontaneously undergo the S to R transition without having been cleaved by a protease, to form a conformation termed the latent state. Latent serpins are unable to interact with proteases and so are no longer protease inhibitors. The conformational change to latency is not exactly the same as the S to R transition of a cleaved serpin. Since the RCL is still intact, the first strand of the C-sheet has to peel off to allow full RCL insertion.[68]
Regulation of the latency transition can act as a control mechanism in some serpins, such as PAI-1. Although PAI-1 is produced in the inhibitory S conformation, it "auto-inactivates" by changing to the latent state unless it is bound to the cofactor vitronectin.[68] Similarly, antithrombin can also spontaneously convert to the latent state, as an additional modulation mechanism to its allosteric activation by heparin.[69] Finally, the N-terminus of tengpin, a serpin from Thermoanaerobacter tengcongensis, is required to lock the molecule in the native inhibitory state. Disruption of interactions made by the N-terminal region results in spontaneous conformational change of this serpin to the latent conformation.[70][71]
Conformational change in non-inhibitory functions
[edit]Certain non-inhibitory serpins also use the serpin conformational change as part of their function. For example, the native (S) form of thyroxine-binding globulin has high affinity for thyroxine, whereas the cleaved (R) form has low affinity. Similarly, transcortin has higher affinity for cortisol when in its native (S) state, than its cleaved (R) state. Thus, in these serpins, RCL cleavage and the S to R transition has been commandeered to allow for ligand release, rather than protease inhibition.[54][55][72]
In some serpins, the S to R transition can activate cell signalling events. In these cases, a serpin that has formed a complex with its target protease, is then recognised by a receptor. The binding event then leads to downstream signalling by the receptor.[73] The S to R transition is therefore used to alert cells to the presence of protease activity.[73] This differs from the usual mechanism whereby serpins affect signalling simply by inhibiting proteases involved in a signalling cascade.[47][48]
Degradation
[edit]When a serpin inhibits a target protease, it forms a permanent complex, which needs to be disposed of. For extracellular serpins, the final serpin-enzyme complexes are rapidly cleared from circulation. One mechanism by which this occurs in mammals is via the low-density lipoprotein receptor-related protein (LRP), which binds to inhibitory complexes made by antithrombin, PA1-1, and neuroserpin, causing cellular uptake.[73][74] Similarly, the Drosophila necrotic serpin is degraded in the lysosome after being trafficked into the cell by the Lipophorin Receptor-1 (homologous to the mammalian LDL receptor family).[75]
Disease and serpinopathies
[edit]Serpins are involved in a wide array of physiological functions, and so mutations in genes encoding them can cause a range of diseases. Mutations that change the activity, specificity or aggregation properties of serpins all affect how they function. The majority of serpin-related diseases are the result of serpin polymerisation into aggregates, though several other types of disease-linked mutations also occur.[8][76] The disorder alpha-1 antitrypsin deficiency is one of the most common hereditary diseases.[11][77]
Inactivity or absence
[edit]
Since the stressed serpin fold is high-energy, mutations can cause them to incorrectly change into their lower-energy conformations (e.g. relaxed or latent) before they have correctly performed their inhibitory role.[10]
Mutations that affect the rate or the extent of RCL insertion into the A-sheet can cause the serpin to undergo its S to R conformational change before having engaged a protease. Since a serpin can only make this conformational change once, the resulting misfired serpin is inactive and unable to properly control its target protease.[10][78] Similarly, mutations that promote inappropriate transition to the monomeric latent state cause disease by reducing the amount of active inhibitory serpin. For example, the disease-linked antithrombin variants wibble and wobble,[79] both promote formation of the latent state.
The structure of the disease-linked mutant of antichymotrypsin (L55P) revealed another, inactive "δ-conformation". In the δ-conformation, four residues of the RCL are inserted into the top of β-sheet A. The bottom half of the sheet is filled as a result of one of the α-helices (the F-helix) partially switching to a β-strand conformation, completing the β-sheet hydrogen bonding.[80] It is unclear whether other serpins can adopt this conformer, and whether this conformation has a functional role, but it is speculated that the δ-conformation may be adopted by Thyroxine-binding globulin during thyroxine release.[55] The non-inhibitory proteins related to serpins can also cause diseases when mutated. For example, mutations in SERPINF1 cause osteogenesis imperfecta type VI in humans.[81]
In the absence of a required serpin, the protease that it normally would regulate is over-active, leading to pathologies.[10] Consequently, simple deficiency of a serpin (e.g. a null mutation) can result in disease.[82] Gene knockouts, particularly in mice, are used experimentally to determine the normal functions of serpins by the effect of their absence.[83]
Specificity change
[edit]In some rare cases, a single amino acid change in a serpin's RCL alters its specificity to target the wrong protease. For example, the Antitrypsin-Pittsburgh mutation (M358R) causes the α1-antitrypsin serpin to inhibit thrombin, causing a bleeding disorder.[28]
Polymerisation and aggregation
[edit]The majority of serpin diseases are due to protein aggregation and are termed "serpinopathies".[12][80] Serpins are vulnerable to disease-causing mutations that promote formation of misfolded polymers due to their inherently unstable structures.[80] Well-characterised serpinopathies include α1-antitrypsin deficiency (alpha-1), which may cause familial emphysema, and sometimes liver cirrhosis, certain familial forms of thrombosis related to antithrombin deficiency, types 1 and 2 hereditary angioedema (HAE) related to deficiency of C1-inhibitor, and familial encephalopathy with neuroserpin inclusion bodies (FENIB; a rare type of dementia caused by neuroserpin polymerisation).[11][12][84]
Each monomer of the serpin aggregate exists in the inactive, relaxed conformation (with the RCL inserted into the A-sheet). The polymers are therefore hyperstable to temperature and unable to inhibit proteases. Serpinopathies therefore cause pathologies similarly to other proteopathies (e.g. prion diseases) via two main mechanisms.[11][12] First, the lack of active serpin results in uncontrolled protease activity and tissue destruction. Second, the hyperstable polymers themselves clog up the endoplasmic reticulum of cells that synthesize serpins, eventually resulting in cell death and tissue damage. In the case of antitrypsin deficiency, antitrypsin polymers cause the death of liver cells, sometimes resulting in liver damage and cirrhosis. Within the cell, serpin polymers are slowly removed via degradation in the endoplasmic reticulum.[85] However, the details of how serpin polymers cause cell death remains to be fully understood.[11]
Physiological serpin polymers are thought to form via domain swapping events, where a segment of one serpin protein inserts into another.[86] Domain-swaps occur when mutations or environmental factors interfere with the final stages of serpin folding to the native state, causing high-energy intermediates to misfold.[87] Both dimer and trimer domain-swap structures have been solved. In the dimer (of antithrombin), the RCL and part of the A-sheet incorporates into the A-sheet of another serpin molecule.[86] The domain-swapped trimer (of antitrypsin) forms via the exchange of an entirely different region of the structure, the B-sheet (with each molecule's RCL inserted into its own A-sheet).[88] It has also been proposed that serpins may form domain-swaps by inserting the RCL of one protein into the A-sheet of another (A-sheet polymerisation).[84][89] These domain-swapped dimer and trimer structures are thought to be the building blocks of the disease-causing polymer aggregates, but the exact mechanism is still unclear.[86][87][88][90]
Therapeutic strategies
[edit]Several therapeutic approaches are in use or under investigation to treat the most common serpinopathy: antitrypsin deficiency.[11] Antitrypsin augmentation therapy is approved for severe antitrypsin deficiency-related emphysema.[91] In this therapy, antitrypsin is purified from the plasma of blood donors and administered intravenously (first marketed as Prolastin).[11][92] To treat severe antitrypsin deficiency-related disease, lung and liver transplantation has proven effective.[11][93] In animal models, gene targeting in induced pluripotent stem cells has been successfully used to correct an antitrypsin polymerisation defect and to restore the ability of the mammalian liver to secrete active antitrypsin.[94] Small molecules have also been developed that block antitrypsin polymerisation in vitro.[95][96]
Evolution
[edit]Serpins are the most widely distributed and largest superfamily of protease inhibitors.[1][40] They were initially believed to be restricted to eukaryote organisms, but have since been found in bacteria, archaea and some viruses.[36][37][97] It remains unclear whether prokaryote genes are the descendants of an ancestral prokaryotic serpin or the product of horizontal gene transfer from eukaryotes. Most intracellular serpins belong to a single phylogenetic clade, whether they come from plants or animals, indicating that the intracellular and extracellular serpins may have diverged before the plants and animals.[98] Exceptions include the intracellular heat shock serpin HSP47, which is a chaperone essential for proper folding of collagen, and cycles between the cis-Golgi and the endoplasmic reticulum.[57]
Protease-inhibition is thought to be the ancestral function, with non-inhibitory members the results of evolutionary neofunctionalisation of the structure. The S to R conformational change has also been adapted by some binding serpins to regulate affinity for their targets.[55]
Distribution
[edit]Animal
[edit]Human
[edit]The human genome encodes 16 serpin clades, termed serpinA through serpinP, including 29 inhibitory and 7 non-inhibitory serpin proteins.[9][83] The human serpin naming system is based upon a phylogenetic analysis of approximately 500 serpins from 2001, with proteins named serpinXY, where X is the clade of the protein and Y the number of the protein within that clade.[1][36][83] The functions of human serpins have been determined by a combination of biochemical studies, human genetic disorders, and knockout mouse models.[83]
| Gene name | Common Name | Localisation | Function / Activity[9][83] | Effect of deficiency[9][83] | Human disease | Chromosomal location | Protein structure |
|---|---|---|---|---|---|---|---|
| SERPINA1 | α1-antitrypsin | Extracellular | Inhibitor of human neutrophil elastase.[99] The C-terminal fragment of cleaved SERPINA1 may inhibit HIV-1 infection.[100] | Deficiency results in emphysema, polymerisation results in cirrhosis (serpinopathy).[11][101] | 14q32.1 | 1QLP, 7API, 1D5S | |
| SERPINA2 | Antitrypsin-related protein | Extracellular | Possible pseudogene.[102] | 14q32.1 | |||
| SERPINA3 | α1-antichymotrypsin | Extracellular | Inhibitor of cathepsin G.[103] Additional roles in chromatin condensation in hepatic cells.[104] | Mis-regulation results in Alzheimer's disease (serpinopathy).[105] | 14q32.1 | 1YXA, 2ACH | |
| SERPINA4 | Kallistatin | Extracellular | Inhibitor of kallikrein, regulator of vascular function.[106][107] | Depletion in hypertensive rats exacerbates renal and cardiovascular injury.[108] | 14q32.1 | ||
| SERPINA5 | Protein C inhibitor | Extracellular | Inhibitor of active protein C.[109] Intracellular role in preventing phagocytosis of bacteria.[110] | Knockout in male mice causes infertility.[111] Accumulation occurs in chronic active plaques in multiple sclerosis.[112] | 14q32.1 | 2OL2, 3B9F | |
| SERPINA6 | Transcortin | Extracellular | Non-inhibitory. Cortisol binding.[54] | Deficiency associated with chronic fatigue.[113] | 14q32.1 | 2V6D, 2VDX, 2VDY | |
| SERPINA7 | Thyroxine-binding globulin | Extracellular | Non-inhibitory. Thyroxine binding.[55] | Deficiency causes hypothyroidism.[114][115] | Xq22.2 | 2CEO, 2RIV, 2RIW | |
| SERPINA8 | Angiotensinogen | Extracellular | Non-inhibitory, cleavage by renin results in release of angiotensin I.[116] | Knockout in mice causes hypotension.[117] | Variants linked to hypertension.[118][119][120] | 1q42-q43 | 2X0B, 2WXW, 2WXX, 2WXY, 2WXZ, 2WY0, 2WY1 |
| SERPINA9 | Centerin / GCET1 | Extracellular | Inhibitory, maintenance of naive B cells.[121][122] | Strongly expressed in most B-cell lymphomas.[123][124] | 14q32.1 | ||
| SERPINA10 | Protein Z-related protease inhibitor | Extracellular | Binds protein Z and inactivates factor Xa and factor XIa.[125] | 14q32.1 | 3F1S, 3H5C | ||
| SERPINA11 | – | Probably extracellular | Unknown | 14q32.13 | |||
| SERPINA12 | Vaspin | Extracellular | Inhibitor of Kallikrein-7. Insulin-sensitizing adipocytokine.[126] | High plasma levels associated with type II diabetes.[127] | 14q32.1 | 4IF8 | |
| SERPINA13 | – | Probably extracellular | Unknown | 14q32 | |||
| SERPINB1 | Monocyte neutrophil elastase inhibitor | Intracellular | Inhibitor of neutrophil elastase.[128] | Knockout in mice causes neutrophil survival defect and immune deficiency.[129] | 6p25 | 1HLE | |
| SERPINB2 | Plasminogen activator inhibitor-2 | Intracellular/extracellular | Inhibitor of extracellular uPA. Intracellular function unclear, but may protect against viral infection.[130] | Deficiency in mice reduces immune response to nematode infection.[131] Knockout in mice causes no obvious phenotype.[132] | 18q21.3 | 1BY7 | |
| SERPINB3 | Squamous cell carcinoma antigen-1 (SCCA-1) | Intracellular | Inhibitor of papain-like cysteine proteases[44] and cathepsins K, L and S.[133][134] | Knockout in mice of Serpinb3a (the murine homolog of both human SERPINB3 and SERPINB4) have reduced mucus production in a murine model of asthma.[135] | 18q21.3 | 2ZV6 | |
| SERPINB4 | Squamous cell carcinoma antigen-2 (SCCA-2) | Intracellular | Inhibitor of chymotrypsin-like serine proteases, cathepsin G and chymase.[134][136] | Knockout in mice of Serpinb3a (the murine homolog of both human SERPINB3 and SERPINB4) have reduced mucus production in a murine model of asthma.[135] | 18q21.3 | ||
| SERPINB5 | Maspin | Intracellular | Non-inhibitory, function unclear[137][138][139] (see also maspin) | Knockout in mice originally reported as lethal,[140] but subsequently shown to have no obvious phenotype.[139] Expression may be a prognostic indicator that reflects expression of a neighbouring tumour suppressor gene (the phosphatase PHLPP1).[139] | 18q21.3 | 1WZ9 | |
| SERPINB6 | PI-6 | Intracellular | Inhibitor of cathepsin G.[141] | Knockout in mice causes hearing loss[142] and mild neutropenia.[143] | Deficiency associated with hearing loss.[144] | 6p25 | |
| SERPINB7 | Megsin | Intracellular | Involved in megakaryocyte maturation.[145] | Over-expression in mice causes kidney disease.[146] Knockout in mice does not cause histological abnormalities.[146] | Mutations associated with Nagashima-type Palmoplantar Keratosis.[147] | 18q21.3 | |
| SERPINB8 | PI-8 | Intracellular | Possible inhibitor of furin.[148] | 18q21.3 | |||
| SERPINB9 | PI-9 | Intracellular | Inhibitor of the cytotoxic granule protease granzyme B.[149] | Knockout in mice causes immune dysfunction.[150][151] | 6p25 | ||
| SERPINB10 | Bomapin | Intracellular | Unknown[152] | Knockout in mice causes no obvious phenotype (C57/BL6; lab strain BC069938). | 18q21.3 | ||
| SERPINB11 | Intracellular | Unknown[153] | Murine Serpinb11 is an active inhibitor whereas the human orthalogue is inactive.[153] Deficiency in ponies is associated with hoof wall separation disease.[154] | 18q21.3 | |||
| SERPINB12 | Yukopin | Intracellular | Unknown[155] | 18q21.3 | |||
| SERPINB13 | Hurpin/Headpin | Intracellular | Inhibitor of papain-like cysteine proteases.[156] | 18q21.3 | |||
| SERPINC1 | Antithrombin | Extracellular | Inhibitor of coagulation, specifically factor X, factor IX and thrombin.[34] | Knockouts in mice are lethal.[157] | Deficiency results in thrombosis and other clotting disorders (serpinopathy).[158][159] | 1q23-q21 | 2ANT, 2ZNH, 1AZX, 1TB6, 2GD4, 1T1F |
| SERPIND1 | Heparin cofactor II | Extracellular | Inhibitor of thrombin.[160] | Knockouts in mice are lethal.[161] | 22q11 | 1JMJ, 1JMO | |
| SERPINE1 | Plasminogen activator inhibitor 1 | Extracellular | Inhibitor of thrombin, uPA and TPa.[162] | 7q21.3-q22 | 1DVN, 1OC0 | ||
| SERPINE2 | Glia derived nexin / Protease nexin I | Extracellular | Inhibitor of uPA and tPA.[163] | Abnormal expression leads to male infertility.[164] Knockout in mice causes epilepsy.[165] | 2q33-q35 | 4DY0 | |
| SERPINF1 | Pigment epithelium derived factor | Extracellular | Non-inhibitory, potent anti-angiogenic molecule.[166] PEDF has been reported to bind the glycosaminoglycan hyaluronan.[167] | Knockout in mice affects the vasculature and mass of the pancreas and the prostate.[166] Promotes Notch–dependent renewal of adult periventricular neural stem cells.[168] Mutations in humans cause osteogenesis imperfecta type VI.[81] | 17p13.3 | 1IMV | |
| SERPINF2 | α2-antiplasmin | Extracellular | Inhibitor of plasmin, inhibitor of fibrinolysis.[169] | Knockouts in mice show increased mice show increased fibrinolysis but no bleeding disorder.[170] | Deficiency causes a rare bleeding disorder.[171][172] | 17pter-p12 | 2R9Y |
| SERPING1 | Complement 1-inhibitor | Extracellular | Inhibitor of C1 esterase.[173] | Several polymorphisms associated with macular degeneration[174] and hereditary angeoedema.[175] | 11q11-q13.1 | 2OAY | |
| SERPINH1 | 47 kDa Heat shock protein (HSP47) | Intracellular | Non-inhibitory, molecular chaperone in collagen folding.[57] | Knockouts in mice are lethal.[176] | Mutation in humans causes severe osteogenesis imperfecta.[177][178] | 11p15 | 4AXY |
| SERPINI1 | Neuroserpin | Extracellular | Inhibitor of tPA, uPA and plasmin.[179] | Mutation causes FENIB dementia (serpinopathy).[180][181] | 3q26 | 1JJO, 3FGQ, 3F5N, 3F02 | |
| SERPINI2 | Pancpin | Extracellular | Unknown[182] | Deficiency in mice causes pancreatic insufficiency via acinar cell loss.[183] | 3q26 |
Specialised mammalian serpins
[edit]Many mammalian serpins have been identified that share no obvious orthology with a human serpin counterpart. Examples include numerous rodent serpins (particularly some of the murine intracellular serpins) as well as the uterine serpins. The term uterine serpin refers to members of the serpin A clade that are encoded by the SERPINA14 gene. Uterine serpins are produced by the endometrium of a restricted group of mammals in the Laurasiatheria clade under the influence of progesterone or estrogen.[184] They are probably not functional proteinase inhibitors and may function during pregnancy to inhibit maternal immune responses against the conceptus or to participate in transplacental transport.[185]
Insect
[edit]The Drosophila melanogaster genome contains 29 serpin encoding genes. Amino acid sequence analysis has placed 14 of these serpins in serpin clade Q and three in serpin clade K with the remaining twelve classified as orphan serpins not belonging to any clade.[186] The clade classification system is difficult to use for Drosophila serpins and instead a nomenclature system has been adopted that is based on the position of serpin genes on the Drosophila chromosomes. Thirteen of the Drosophila serpins occur as isolated genes in the genome (including Serpin-27A, see below), with the remaining 16 organised into five gene clusters that occur at chromosome positions 28D (2 serpins), 42D (5 serpins), 43A (4 serpins), 77B (3 serpins) and 88E (2 serpins).[186][187][188]
Studies on Drosophila serpins reveal that Serpin-27A inhibits the Easter protease (the final protease in the Nudel, Gastrulation Defective, Snake and Easter proteolytic cascade) and thus controls dorsoventral patterning. Easter functions to cleave Spätzle (a chemokine-type ligand), which results in toll-mediated signaling. As well as its central role in embryonic patterning, toll signaling is also important for the innate immune response in insects. Accordingly, serpin-27A also functions to control the insect immune response.[48][189][190] In Tenebrio molitor (a large beetle), a protein (SPN93) comprising two discrete tandem serpin domains functions to regulate the toll proteolytic cascade.[191]
Nematode
[edit]The genome of the nematode worm C. elegans contains 9 serpins, all of which lack signal sequences and so are likely intracellular.[192] However, only 5 of these serpins appear to function as protease inhibitors.[192] One, SRP-6, performs a protective function and guards against stress-induced calpain-associated lysosomal disruption. Further, SRP-6 inhibits lysosomal cysteine proteases released after lysosomal rupture. Accordingly, worms lacking SRP-6 are sensitive to stress. Most notably, SRP-6 knockout worms die when placed in water (the hypo-osmotic stress lethal phenotype or Osl). It has therefore been suggested that lysosomes play a general and controllable role in determining cell fate.[193]
Plant
[edit]Plant serpins were amongst the first members of the superfamily that were identified.[194] The serpin barley protein Z is highly abundant in barley grain, and one of the major protein components in beer. The genome of the model plant, Arabidopsis thaliana contain 18 serpin-like genes, although only 8 of these are full-length serpin sequences.
Plant serpins are potent inhibitors of mammalian chymotrypsin-like serine proteases in vitro, the best-studied example being barley serpin Zx (BSZx), which is able to inhibit trypsin and chymotrypsin as well as several blood coagulation factors.[195] However, close relatives of chymotrypsin-like serine proteases are absent in plants. The RCL of several serpins from wheat grain and rye contain poly-Q repeat sequences similar to those present in the prolamin storage proteins of the endosperm.[196][197] It has therefore been suggested that plant serpins may function to inhibit proteases from insects or microbes that would otherwise digest grain storage proteins. In support of this hypothesis, specific plant serpins have been identified in the phloem sap of pumpkin (CmPS-1)[198] and cucumber plants.[199][200] Although an inverse correlation between up-regulation of CmPS-1 expression and aphid survival was observed, in vitro feeding experiments revealed that recombinant CmPS-1 did not appear to affect insect survival.[198]
Alternative roles and protease targets for plant serpins have been proposed. The Arabidopsis serpin, AtSerpin1 (At1g47710; 3LE2), mediates set-point control over programmed cell death by targeting the 'Responsive to Desiccation-21' (RD21) papain-like cysteine protease.[53][201] AtSerpin1 also inhibits metacaspase-like proteases in vitro.[52] Two other Arabidopsis serpins, AtSRP2 (At2g14540) and AtSRP3 (At1g64030) appear to be involved in responses to DNA damage.[202]
Fungal
[edit]A single fungal serpin has been characterized to date: celpin from Piromyces spp. strain E2. Piromyces is a genus of anaerobic fungi found in the gut of ruminants and is important for digesting plant material. Celpin is predicted to be inhibitory and contains two N-terminal dockerin domains in addition to its serpin domain. Dockerins are commonly found in proteins that localise to the fungal cellulosome, a large extracellular multiprotein complex that breaks down cellulose.[38] It is therefore suggested that celpin may protect the cellulosome against plant proteases. Certain bacterial serpins similarly localize to the cellulosome.[203]
Prokaryotic
[edit]Predicted serpin genes are sporadically distributed in prokaryotes. In vitro studies on some of these molecules have revealed that they are able to inhibit proteases, and it is suggested that they function as inhibitors in vivo. Several prokaryote serpins are found in extremophiles. Accordingly, and in contrast to mammalian serpins, these molecules possess elevated resistance to heat denaturation.[204][205] The precise role of most bacterial serpins remains obscure, although Clostridium thermocellum serpin localises to the cellulosome. It is suggested that the role of cellulosome-associated serpins may be to prevent unwanted protease activity against the cellulosome.[203]
Viral
[edit]Serpins are also expressed by viruses as a way to evade the host's immune defense.[206] In particular, serpins expressed by pox viruses, including cow pox (vaccinia) and rabbit pox (myxoma), are of interest because of their potential use as novel therapeutics for immune and inflammatory disorders as well as transplant therapy.[207][208] Serp1 suppresses the TLR-mediated innate immune response and allows indefinite cardiac allograft survival in rats.[207][209] Crma and Serp2 are both cross-class inhibitors and target both serine (granzyme B; albeit weakly) and cysteine proteases (caspase 1 and caspase 8).[210][211] In comparison to their mammalian counterparts, viral serpins contain significant deletions of elements of secondary structure. Specifically, crmA lacks the D-helix as well as significant portions of the A- and E-helices.[212]
References
[edit]- ^ a b c d e Silverman GA, Bird PI, Carrell RW, Church FC, Coughlin PB, Gettins PG, Irving JA, Lomas DA, Luke CJ, Moyer RW, Pemberton PA, Remold-O'Donnell E, Salvesen GS, Travis J, Whisstock JC (September 2001). "The serpins are an expanding superfamily of structurally similar but functionally diverse proteins. Evolution, mechanism of inhibition, novel functions, and a revised nomenclature". The Journal of Biological Chemistry. 276 (36): 33293–33296. doi:10.1074/jbc.R100016200. PMID 11435447.
- ^ Spence MA, Mortimer MD, Buckle AM, Minh BQ, Jackson CJ (June 2021). Echave J (ed.). "A Comprehensive Phylogenetic Analysis of the Serpin Superfamily". Molecular Biology and Evolution. 38 (7): 2915–2929. doi:10.1093/molbev/msab081. PMC 8233489. PMID 33744972.
- ^ Carrell RW, Boswell DR (1986). "Serpins: the superfamily of plasma serine proteinase inhibitors". In Barrett AJ, Salvesen G (eds.). Proteinase Inhibitors. Research monographs in cell and tissue physiology. Vol. 12. Amsterdam: Elsevier Science Publishers BV. pp. 403–420. ISBN 0-444-80763-2.
- ^ Silverman GA, Whisstock JC, Bottomley SP, Huntington JA, Kaiserman D, Luke CJ, Pak SC, Reichhart JM, Bird PI (August 2010). "Serpins flex their muscle: I. Putting the clamps on proteolysis in diverse biological systems". The Journal of Biological Chemistry. 285 (32): 24299–24305. doi:10.1074/jbc.R110.112771. PMC 2915665. PMID 20498369.
- ^ Whisstock JC, Silverman GA, Bird PI, Bottomley SP, Kaiserman D, Luke CJ, Pak SC, Reichhart JM, Huntington JA (August 2010). "Serpins flex their muscle: II. Structural insights into target peptidase recognition, polymerization, and transport functions". The Journal of Biological Chemistry. 285 (32): 24307–24312. doi:10.1074/jbc.R110.141408. PMC 2915666. PMID 20498368.
- ^ a b c d Huntington JA, Read RJ, Carrell RW (October 2000). "Structure of a serpin-protease complex shows inhibition by deformation". Nature. 407 (6806): 923–926. Bibcode:2000Natur.407..923H. doi:10.1038/35038119. PMID 11057674. S2CID 205009937.
- ^ a b c d e f g Gettins PG (December 2002). "Serpin structure, mechanism, and function". Chemical Reviews. 102 (12): 4751–4804. doi:10.1021/cr010170. PMID 12475206.
- ^ a b c d e f g Whisstock JC, Bottomley SP (December 2006). "Molecular gymnastics: serpin structure, folding and misfolding". Current Opinion in Structural Biology. 16 (6): 761–768. doi:10.1016/j.sbi.2006.10.005. PMID 17079131.
- ^ a b c d e f Law RH, Zhang Q, McGowan S, Buckle AM, Silverman GA, Wong W, Rosado CJ, Langendorf CG, Pike RN, Bird PI, Whisstock JC (2006). "An overview of the serpin superfamily". Genome Biology. 7 (5): 216. doi:10.1186/gb-2006-7-5-216. PMC 1779521. PMID 16737556.
- ^ a b c d e Stein PE, Carrell RW (February 1995). "What do dysfunctional serpins tell us about molecular mobility and disease?". Nature Structural Biology. 2 (2): 96–113. doi:10.1038/nsb0295-96. PMID 7749926. S2CID 21223825.
- ^ a b c d e f g h i Janciauskiene SM, Bals R, Koczulla R, Vogelmeier C, Köhnlein T, Welte T (August 2011). "The discovery of α1-antitrypsin and its role in health and disease". Respiratory Medicine. 105 (8): 1129–1139. doi:10.1016/j.rmed.2011.02.002. PMID 21367592.
- ^ a b c d Carrell RW, Lomas DA (July 1997). "Conformational disease". Lancet. 350 (9071): 134–138. doi:10.1016/S0140-6736(97)02073-4. PMID 9228977. S2CID 39124185.
- ^ Fermi C, Pernossi L (December 1894). "Untersuchungen uber die enzyme, Vergleichende Studie" [Studies on the enzyme, Comparative study] (PDF). Zeitschrift für Hygiene und Infektionskrankheiten (in German) (18): 83–89. doi:10.1007/BF02216836. S2CID 24373770.
- ^ Schultze HU, Göllner I, Heide K, Schönenberger M, Schwick G (August 1955). "Zur Kenntnis der alpha-globulin des menschlichen normal serums" [For knowledge of the alpha - globulin of human normal serums]. Zeitschrift für Naturforschung B (in German). 10 (8): 463. doi:10.1515/znb-1955-0810. S2CID 95960716.
- ^ Petersen TE, Dudeck-Wojciechowska G, Sottrup-Jensen L, Magnusson S (1979). "Primary structure of antithrombin III (heparin cofactor): partial homology between alpha-1-antitrypsin and antithrombin III". In Collen D, Wiman B, Verstraete M (eds.). The Physiological Inhibitors of Coagulation and Fibrinolysis. Amsterdam: Elsevier. pp. 43–54.
- ^ Carrell R, Owen M, Brennan S, Vaughan L (December 1979). "Carboxy terminal fragment of human alpha-1-antitrypsin from hydroxylamine cleavage: homology with antithrombin III". Biochemical and Biophysical Research Communications. 91 (3): 1032–1037. doi:10.1016/0006-291X(79)91983-1. PMID 316698.
- ^ Hunt LT, Dayhoff MO (July 1980). "A surprising new protein superfamily containing ovalbumin, antithrombin-III, and alpha 1-proteinase inhibitor". Biochemical and Biophysical Research Communications. 95 (2): 864–871. doi:10.1016/0006-291X(80)90867-0. PMID 6968211.
- ^ Carrell RW, Jeppsson JO, Laurell CB, Brennan SO, Owen MC, Vaughan L, Boswell DR (July 1982). "Structure and variation of human alpha 1-antitrypsin". Nature. 298 (5872): 329–334. doi:10.1016/0968-0004(85)90011-8. PMC 7172600. PMID 7045697.
- ^ Laurell CB, Eriksson S (March 2013). "The electrophoretic α1-globulin pattern of serum in α1-antitrypsin deficiency. 1963". Copd. 10 (Suppl 1): 3–8. doi:10.3109/15412555.2013.771956. PMID 23527532. S2CID 36366089.
- ^ Sharp HL, Bridges RA, Krivit W, Freier EF (June 1969). "Cirrhosis associated with alpha-1-antitrypsin deficiency: a previously unrecognized inherited disorder". The Journal of Laboratory and Clinical Medicine. 73 (6): 934–939. PMID 4182334.
- ^ "Alpha-1-antitrypsin: molecular abnormality of S variant". British Medical Journal. 1 (6002): 130–131. January 1976. doi:10.1136/bmj.1.6002.130-a. PMC 1638590. PMID 1082356.
- ^ Jeppsson JO (June 1976). "Amino acid substitution Glu leads to Lys alpha1-antitrypsin PiZ". FEBS Letters. 65 (2): 195–197. doi:10.1016/0014-5793(76)80478-4. PMID 1084290. S2CID 84576569.
- ^ Carrell RW, Jeppsson JO, Laurell CB, Brennan SO, Owen MC, Vaughan L, Boswell DR (July 1982). "Structure and variation of human alpha 1-antitrypsin". Nature. 298 (5872): 329–334. Bibcode:1982Natur.298..329C. doi:10.1038/298329a0. PMID 7045697. S2CID 11904305.
- ^ Carrell RW, Boswell DR, Brennan SO, Owen MC (March 1980). "Active site of alpha 1-antitrypsin: homologous site in antithrombin-III". Biochemical and Biophysical Research Communications. 93 (2): 399–402. doi:10.1016/0006-291X(80)91090-6. PMID 6966929.
- ^ Johnson D, Travis J (October 1978). "Structural evidence for methionine at the reactive site of human alpha-1-proteinase inhibitor". The Journal of Biological Chemistry. 253 (20): 7142–7144. doi:10.1016/S0021-9258(17)34475-7. PMID 701239.
- ^ Jörnvall H, Fish WW, Björk I (October 1979). "The thrombin cleavage site in bovine antithrombin". FEBS Letters. 106 (2): 358–362. Bibcode:1979FEBSL.106..358J. doi:10.1016/0014-5793(79)80532-3. PMID 499520. S2CID 32540550.
- ^ Egeberg O (June 1965). "Inherited antithrombin deficiency causing thrombophilia". Thrombosis et Diathesis Haemorrhagica. 13 (2): 516–530. doi:10.1055/s-0038-1656297. PMID 14347873. S2CID 42594050.
- ^ a b Owen MC, Brennan SO, Lewis JH, Carrell RW (September 1983). "Mutation of antitrypsin to antithrombin. alpha 1-antitrypsin Pittsburgh (358 Met leads to Arg), a fatal bleeding disorder". The New England Journal of Medicine. 309 (12): 694–698. doi:10.1056/NEJM198309223091203. PMID 6604220.
- ^ a b Loebermann H, Tokuoka R, Deisenhofer J, Huber R (August 1984). "Human alpha 1-proteinase inhibitor. Crystal structure analysis of two crystal modifications, molecular model and preliminary analysis of the implications for function". Journal of Molecular Biology. 177 (3): 531–557. doi:10.1016/0022-2836(84)90298-5. PMID 6332197.
- ^ a b Stein PE, Leslie AG, Finch JT, Turnell WG, McLaughlin PJ, Carrell RW (September 1990). "Crystal structure of ovalbumin as a model for the reactive centre of serpins". Nature. 347 (6288): 99–102. Bibcode:1990Natur.347...99S. doi:10.1038/347099a0. PMID 2395463. S2CID 4342263.
- ^ a b Carrell RW, Evans DL, Stein PE (October 1991). "Mobile reactive centre of serpins and the control of thrombosis". Nature. 353 (6344): 576–578. Bibcode:1991Natur.353..576C. doi:10.1038/353576a0. PMID 1922367. S2CID 4361304.
- ^ Mottonen J, Strand A, Symersky J, Sweet RM, Danley DE, Geoghegan KF, Gerard RD, Goldsmith EJ (January 1992). "Structural basis of latency in plasminogen activator inhibitor-1". Nature. 355 (6357): 270–273. Bibcode:1992Natur.355..270M. doi:10.1038/355270a0. PMID 1731226. S2CID 4365370.
- ^ Austin RC, Rachubinski RA, Ofosu FA, Blajchman MA (May 1991). "Antithrombin-III-Hamilton, Ala 382 to Thr: an antithrombin-III variant that acts as a substrate but not an inhibitor of alpha-thrombin and factor Xa". Blood. 77 (10): 2185–2189. doi:10.1182/blood.V77.10.2185.2185. PMID 2029579.
- ^ a b c Huntington JA (August 2006). "Shape-shifting serpins--advantages of a mobile mechanism". Trends in Biochemical Sciences. 31 (8): 427–435. doi:10.1016/j.tibs.2006.06.005. PMID 16820297.
- ^ Carrell RW, Read RJ (February 2017). "How serpins transport hormones and regulate their release". Seminars in Cell & Developmental Biology. 62: 133–141. doi:10.1016/j.semcdb.2016.12.007. PMID 28027946.
- ^ a b c Irving JA, Pike RN, Lesk AM, Whisstock JC (December 2000). "Phylogeny of the serpin superfamily: implications of patterns of amino acid conservation for structure and function". Genome Research. 10 (12): 1845–1864. doi:10.1101/gr.GR-1478R. PMID 11116082.
- ^ a b Irving JA, Steenbakkers PJ, Lesk AM, Op den Camp HJ, Pike RN, Whisstock JC (November 2002). "Serpins in prokaryotes". Molecular Biology and Evolution. 19 (11): 1881–1890. doi:10.1093/oxfordjournals.molbev.a004012. PMID 12411597.
- ^ a b Steenbakkers PJ, Irving JA, Harhangi HR, Swinkels WJ, Akhmanova A, Dijkerman R, Jetten MS, van der Drift C, Whisstock JC, Op den Camp HJ (August 2008). "A serpin in the cellulosome of the anaerobic fungus Piromyces sp. strain E2". Mycological Research. 112 (Pt 8): 999–1006. doi:10.1016/j.mycres.2008.01.021. hdl:2066/72679. PMID 18539447.
- ^ Huber R, Carrell RW (November 1989). "Implications of the three-dimensional structure of alpha 1-antitrypsin for structure and function of serpins". Biochemistry. 28 (23): 8951–8966. doi:10.1021/bi00449a001. PMID 2690952.
- ^ a b Rawlings ND, Tolle DP, Barrett AJ (March 2004). "Evolutionary families of peptidase inhibitors". The Biochemical Journal. 378 (Pt 3): 705–716. doi:10.1042/BJ20031825. PMC 1224039. PMID 14705960.
- ^ Barrett AJ, Rawlings ND (April 1995). "Families and clans of serine peptidases". Archives of Biochemistry and Biophysics. 318 (2): 247–250. doi:10.1006/abbi.1995.1227. PMID 7733651.
- ^ Barrett AJ, Rawlings ND (May 2001). "Evolutionary lines of cysteine peptidases". Biological Chemistry. 382 (5): 727–733. doi:10.1515/BC.2001.088. PMID 11517925. S2CID 37306786.
- ^ Irving JA, Pike RN, Dai W, Brömme D, Worrall DM, Silverman GA, Coetzer TH, Dennison C, Bottomley SP, Whisstock JC (April 2002). "Evidence that serpin architecture intrinsically supports papain-like cysteine protease inhibition: engineering alpha(1)-antitrypsin to inhibit cathepsin proteases". Biochemistry. 41 (15): 4998–5004. doi:10.1021/bi0159985. PMID 11939796.
- ^ a b Schick C, Brömme D, Bartuski AJ, Uemura Y, Schechter NM, Silverman GA (November 1998). "The reactive site loop of the serpin SCCA1 is essential for cysteine proteinase inhibition". Proceedings of the National Academy of Sciences of the United States of America. 95 (23): 13465–13470. Bibcode:1998PNAS...9513465S. doi:10.1073/pnas.95.23.13465. PMC 24842. PMID 9811823.
- ^ a b McGowan S, Buckle AM, Irving JA, Ong PC, Bashtannyk-Puhalovich TA, Kan WT, Henderson KN, Bulynko YA, Popova EY, Smith AI, Bottomley SP, Rossjohn J, Grigoryev SA, Pike RN, Whisstock JC (July 2006). "X-ray crystal structure of MENT: evidence for functional loop-sheet polymers in chromatin condensation". The EMBO Journal. 25 (13): 3144–3155. doi:10.1038/sj.emboj.7601201. PMC 1500978. PMID 16810322.
- ^ Ong PC, McGowan S, Pearce MC, Irving JA, Kan WT, Grigoryev SA, Turk B, Silverman GA, Brix K, Bottomley SP, Whisstock JC, Pike RN (December 2007). "DNA accelerates the inhibition of human cathepsin V by serpins". The Journal of Biological Chemistry. 282 (51): 36980–36986. doi:10.1074/jbc.M706991200. PMID 17923478.
- ^ a b Acosta H, Iliev D, Grahn TH, Gouignard N, Maccarana M, Griesbach J, Herzmann S, Sagha M, Climent M, Pera EM (March 2015). "The serpin PN1 is a feedback regulator of FGF signaling in germ layer and primary axis formation". Development. 142 (6): 1146–1158. doi:10.1242/dev.113886. PMID 25758225.
- ^ a b c Hashimoto C, Kim DR, Weiss LA, Miller JW, Morisato D (December 2003). "Spatial regulation of developmental signaling by a serpin". Developmental Cell. 5 (6): 945–950. doi:10.1016/S1534-5807(03)00338-1. PMID 14667416.
- ^ Bird PI (February 1999). "Regulation of pro-apoptotic leucocyte granule serine proteinases by intracellular serpins". Immunology and Cell Biology. 77 (1): 47–57. doi:10.1046/j.1440-1711.1999.00787.x. PMID 10101686. S2CID 44268106.
- ^ Bird CH, Sutton VR, Sun J, Hirst CE, Novak A, Kumar S, Trapani JA, Bird PI (November 1998). "Selective regulation of apoptosis: the cytotoxic lymphocyte serpin proteinase inhibitor 9 protects against granzyme B-mediated apoptosis without perturbing the Fas cell death pathway". Molecular and Cellular Biology. 18 (11): 6387–6398. doi:10.1128/mcb.18.11.6387. PMC 109224. PMID 9774654.
- ^ Ray CA, Black RA, Kronheim SR, Greenstreet TA, Sleath PR, Salvesen GS, Pickup DJ (May 1992). "Viral inhibition of inflammation: cowpox virus encodes an inhibitor of the interleukin-1 beta converting enzyme". Cell. 69 (4): 597–604. doi:10.1016/0092-8674(92)90223-Y. PMID 1339309. S2CID 7398844.
- ^ a b Vercammen D, Belenghi B, van de Cotte B, Beunens T, Gavigan JA, De Rycke R, Brackenier A, Inzé D, Harris JL, Van Breusegem F (December 2006). "Serpin1 of Arabidopsis thaliana is a suicide inhibitor for metacaspase 9". Journal of Molecular Biology. 364 (4): 625–636. doi:10.1016/j.jmb.2006.09.010. PMID 17028019.
- ^ a b Lampl N, Budai-Hadrian O, Davydov O, Joss TV, Harrop SJ, Curmi PM, Roberts TH, Fluhr R (April 2010). "Arabidopsis AtSerpin1, crystal structure and in vivo interaction with its target protease RESPONSIVE TO DESICCATION-21 (RD21)". The Journal of Biological Chemistry. 285 (18): 13550–13560. doi:10.1074/jbc.M109.095075. PMC 2859516. PMID 20181955.
- ^ a b c Klieber MA, Underhill C, Hammond GL, Muller YA (October 2007). "Corticosteroid-binding globulin, a structural basis for steroid transport and proteinase-triggered release". The Journal of Biological Chemistry. 282 (40): 29594–29603. doi:10.1074/jbc.M705014200. PMID 17644521.
- ^ a b c d e Zhou A, Wei Z, Read RJ, Carrell RW (September 2006). "Structural mechanism for the carriage and release of thyroxine in the blood". Proceedings of the National Academy of Sciences of the United States of America. 103 (36): 13321–13326. Bibcode:2006PNAS..10313321Z. doi:10.1073/pnas.0604080103. PMC 1557382. PMID 16938877.
- ^ Huntington JA, Stein PE (May 2001). "Structure and properties of ovalbumin". Journal of Chromatography. B, Biomedical Sciences and Applications. 756 (1–2): 189–198. doi:10.1016/S0378-4347(01)00108-6. PMID 11419711.
- ^ a b c Mala JG, Rose C (November 2010). "Interactions of heat shock protein 47 with collagen and the stress response: an unconventional chaperone model?". Life Sciences. 87 (19–22): 579–586. doi:10.1016/j.lfs.2010.09.024. PMID 20888348.
- ^ Grigoryev SA, Bednar J, Woodcock CL (February 1999). "MENT, a heterochromatin protein that mediates higher order chromatin folding, is a new serpin family member". The Journal of Biological Chemistry. 274 (9): 5626–5636. doi:10.1074/jbc.274.9.5626. PMID 10026180.
- ^ Elliott PR, Lomas DA, Carrell RW, Abrahams JP (August 1996). "Inhibitory conformation of the reactive loop of alpha 1-antitrypsin". Nature Structural Biology. 3 (8): 676–681. doi:10.1038/nsb0896-676. PMID 8756325. S2CID 22976014.
- ^ Horvath AJ, Irving JA, Rossjohn J, Law RH, Bottomley SP, Quinsey NS, Pike RN, Coughlin PB, Whisstock JC (December 2005). "The murine orthologue of human antichymotrypsin: a structural paradigm for clade A3 serpins". The Journal of Biological Chemistry. 280 (52): 43168–43178. doi:10.1074/jbc.M505598200. PMID 16141197.
- ^ Whisstock JC, Skinner R, Carrell RW, Lesk AM (February 2000). "Conformational changes in serpins: I. The native and cleaved conformations of alpha(1)-antitrypsin". Journal of Molecular Biology. 296 (2): 685–699. doi:10.1006/jmbi.1999.3520. PMID 10669617.
- ^ Jin L, Abrahams JP, Skinner R, Petitou M, Pike RN, Carrell RW (December 1997). "The anticoagulant activation of antithrombin by heparin". Proceedings of the National Academy of Sciences of the United States of America. 94 (26): 14683–14688. Bibcode:1997PNAS...9414683J. doi:10.1073/pnas.94.26.14683. PMC 25092. PMID 9405673.
- ^ Whisstock JC, Pike RN, Jin L, Skinner R, Pei XY, Carrell RW, Lesk AM (September 2000). "Conformational changes in serpins: II. The mechanism of activation of antithrombin by heparin". Journal of Molecular Biology. 301 (5): 1287–1305. doi:10.1006/jmbi.2000.3982. PMID 10966821.
- ^ Li W, Johnson DJ, Esmon CT, Huntington JA (September 2004). "Structure of the antithrombin-thrombin-heparin ternary complex reveals the antithrombotic mechanism of heparin". Nature Structural & Molecular Biology. 11 (9): 857–862. doi:10.1038/nsmb811. PMID 15311269. S2CID 28790576.
- ^ Johnson DJ, Li W, Adams TE, Huntington JA (May 2006). "Antithrombin-S195A factor Xa-heparin structure reveals the allosteric mechanism of antithrombin activation". The EMBO Journal. 25 (9): 2029–2037. doi:10.1038/sj.emboj.7601089. PMC 1456925. PMID 16619025.
- ^ Walenga JM, Jeske WP, Samama MM, Frapaise FX, Bick RL, Fareed J (March 2002). "Fondaparinux: a synthetic heparin pentasaccharide as a new antithrombotic agent". Expert Opinion on Investigational Drugs. 11 (3): 397–407. doi:10.1517/13543784.11.3.397. PMID 11866668. S2CID 24796086.
- ^ Petitou M, van Boeckel CA (June 2004). "A synthetic antithrombin III binding pentasaccharide is now a drug! What comes next?". Angewandte Chemie. 43 (24): 3118–3133. doi:10.1002/anie.200300640. PMID 15199558.
- ^ a b Lindahl TL, Sigurdardottir O, Wiman B (September 1989). "Stability of plasminogen activator inhibitor 1 (PAI-1)". Thrombosis and Haemostasis. 62 (2): 748–751. doi:10.1055/s-0038-1646895. PMID 2479113. S2CID 19433778.
- ^ Mushunje A, Evans G, Brennan SO, Carrell RW, Zhou A (December 2004). "Latent antithrombin and its detection, formation and turnover in the circulation". Journal of Thrombosis and Haemostasis. 2 (12): 2170–2177. doi:10.1111/j.1538-7836.2004.01047.x. PMID 15613023. S2CID 43029244.
- ^ Zhang Q, Buckle AM, Law RH, Pearce MC, Cabrita LD, Lloyd GJ, Irving JA, Smith AI, Ruzyla K, Rossjohn J, Bottomley SP, Whisstock JC (July 2007). "The N terminus of the serpin, tengpin, functions to trap the metastable native state". EMBO Reports. 8 (7): 658–663. doi:10.1038/sj.embor.7400986. PMC 1905895. PMID 17557112.
- ^ Zhang Q, Law RH, Bottomley SP, Whisstock JC, Buckle AM (March 2008). "A structural basis for loop C-sheet polymerization in serpins". Journal of Molecular Biology. 376 (5): 1348–1359. doi:10.1016/j.jmb.2007.12.050. PMID 18234218.
- ^ Pemberton PA, Stein PE, Pepys MB, Potter JM, Carrell RW (November 1988). "Hormone binding globulins undergo serpin conformational change in inflammation". Nature. 336 (6196): 257–258. Bibcode:1988Natur.336..257P. doi:10.1038/336257a0. PMID 3143075. S2CID 4326356.
- ^ a b c Cao C, Lawrence DA, Li Y, Von Arnim CA, Herz J, Su EJ, Makarova A, Hyman BT, Strickland DK, Zhang L (May 2006). "Endocytic receptor LRP together with tPA and PAI-1 coordinates Mac-1-dependent macrophage migration". The EMBO Journal. 25 (9): 1860–1870. doi:10.1038/sj.emboj.7601082. PMC 1456942. PMID 16601674.
- ^ Jensen JK, Dolmer K, Gettins PG (July 2009). "Specificity of binding of the low density lipoprotein receptor-related protein to different conformational states of the clade E serpins plasminogen activator inhibitor-1 and proteinase nexin-1". The Journal of Biological Chemistry. 284 (27): 17989–17997. doi:10.1074/jbc.M109.009530. PMC 2709341. PMID 19439404.
- ^ Soukup SF, Culi J, Gubb D (June 2009). Rulifson E (ed.). "Uptake of the necrotic serpin in Drosophila melanogaster via the lipophorin receptor-1". PLOS Genetics. 5 (6): e1000532. doi:10.1371/journal.pgen.1000532. PMC 2694266. PMID 19557185.
- ^ Kaiserman D, Whisstock JC, Bird PI (December 2006). "Mechanisms of serpin dysfunction in disease". Expert Reviews in Molecular Medicine. 8 (31): 1–19. doi:10.1017/S1462399406000184. PMID 17156576. S2CID 20760165.
- ^ de Serres FJ (November 2002). "Worldwide racial and ethnic distribution of alpha1-antitrypsin deficiency: summary of an analysis of published genetic epidemiologic surveys". Chest. 122 (5): 1818–1829. doi:10.1378/chest.122.5.1818. PMID 12426287.
- ^ Hopkins PC, Carrell RW, Stone SR (August 1993). "Effects of mutations in the hinge region of serpins". Biochemistry. 32 (30): 7650–7657. doi:10.1021/bi00081a008. PMID 8347575.
- ^ Beauchamp NJ, Pike RN, Daly M, Butler L, Makris M, Dafforn TR, Zhou A, Fitton HL, Preston FE, Peake IR, Carrell RW (October 1998). "Antithrombins Wibble and Wobble (T85M/K): archetypal conformational diseases with in vivo latent-transition, thrombosis, and heparin activation". Blood. 92 (8): 2696–2706. doi:10.1182/blood.V92.8.2696. PMID 9763552.
- ^ a b c Gooptu B, Hazes B, Chang WS, Dafforn TR, Carrell RW, Read RJ, Lomas DA (January 2000). "Inactive conformation of the serpin alpha(1)-antichymotrypsin indicates two-stage insertion of the reactive loop: implications for inhibitory function and conformational disease". Proceedings of the National Academy of Sciences of the United States of America. 97 (1): 67–72. Bibcode:2000PNAS...97...67G. doi:10.1073/pnas.97.1.67. PMC 26617. PMID 10618372.
- ^ a b Homan EP, Rauch F, Grafe I, Lietman C, Doll JA, Dawson B, Bertin T, Napierala D, Morello R, Gibbs R, White L, Miki R, Cohn DH, Crawford S, Travers R, Glorieux FH, Lee B (December 2011). "Mutations in SERPINF1 cause osteogenesis imperfecta type VI". Journal of Bone and Mineral Research. 26 (12): 2798–2803. doi:10.1002/jbmr.487. PMC 3214246. PMID 21826736.
- ^ Fay WP, Parker AC, Condrey LR, Shapiro AD (July 1997). "Human plasminogen activator inhibitor-1 (PAI-1) deficiency: characterization of a large kindred with a null mutation in the PAI-1 gene". Blood. 90 (1): 204–208. doi:10.1182/blood.V90.1.204. PMID 9207454.
- ^ a b c d e f Heit C, Jackson BC, McAndrews M, Wright MW, Thompson DC, Silverman GA, Nebert DW, Vasiliou V (October 2013). "Update of the human and mouse SERPIN gene superfamily". Human Genomics. 7 (1): 22. doi:10.1186/1479-7364-7-22. PMC 3880077. PMID 24172014.
- ^ a b Lomas DA, Evans DL, Finch JT, Carrell RW (June 1992). "The mechanism of Z alpha 1-antitrypsin accumulation in the liver". Nature. 357 (6379): 605–607. Bibcode:1992Natur.357..605L. doi:10.1038/357605a0. PMID 1608473. S2CID 4359543.
- ^ Kroeger H, Miranda E, MacLeod I, Pérez J, Crowther DC, Marciniak SJ, Lomas DA (August 2009). "Endoplasmic reticulum-associated degradation (ERAD) and autophagy cooperate to degrade polymerogenic mutant serpins". The Journal of Biological Chemistry. 284 (34): 22793–22802. doi:10.1074/jbc.M109.027102. PMC 2755687. PMID 19549782.
- ^ a b c Yamasaki M, Li W, Johnson DJ, Huntington JA (October 2008). "Crystal structure of a stable dimer reveals the molecular basis of serpin polymerization". Nature. 455 (7217): 1255–1258. Bibcode:2008Natur.455.1255Y. doi:10.1038/nature07394. PMID 18923394. S2CID 205215121.
- ^ a b Bottomley SP (September 2011). "The structural diversity in α1-antitrypsin misfolding". EMBO Reports. 12 (10): 983–984. doi:10.1038/embor.2011.187. PMC 3185355. PMID 21921939.
- ^ a b Yamasaki M, Sendall TJ, Pearce MC, Whisstock JC, Huntington JA (September 2011). "Molecular basis of α1-antitrypsin deficiency revealed by the structure of a domain-swapped trimer". EMBO Reports. 12 (10): 1011–1017. doi:10.1038/embor.2011.171. PMC 3185345. PMID 21909074.
- ^ Chang WS, Whisstock J, Hopkins PC, Lesk AM, Carrell RW, Wardell MR (January 1997). "Importance of the release of strand 1C to the polymerization mechanism of inhibitory serpins". Protein Science. 6 (1): 89–98. doi:10.1002/pro.5560060110. PMC 2143506. PMID 9007980.
- ^ Miranda E, Pérez J, Ekeowa UI, Hadzic N, Kalsheker N, Gooptu B, Portmann B, Belorgey D, Hill M, Chambers S, Teckman J, Alexander GJ, Marciniak SJ, Lomas DA (September 2010). "A novel monoclonal antibody to characterize pathogenic polymers in liver disease associated with alpha1-antitrypsin deficiency". Hepatology. 52 (3): 1078–1088. doi:10.1002/hep.23760. PMID 20583215. S2CID 8188156.
- ^ Sandhaus RA (October 2004). "alpha1-Antitrypsin deficiency . 6: new and emerging treatments for alpha1-antitrypsin deficiency". Thorax. 59 (10): 904–909. doi:10.1136/thx.2003.006551. PMC 1746849. PMID 15454659.
- ^ Lewis EC (September 2012). "Expanding the clinical indications for α(1)-antitrypsin therapy". Molecular Medicine. 18 (6): 957–970. doi:10.2119/molmed.2011.00196. PMC 3459478. PMID 22634722.
- ^ Fregonese L, Stolk J (June 2008). "Hereditary alpha-1-antitrypsin deficiency and its clinical consequences". Orphanet Journal of Rare Diseases. 3: 16. doi:10.1186/1750-1172-3-16. PMC 2441617. PMID 18565211.
- ^ Yusa K, Rashid ST, Strick-Marchand H, Varela I, Liu PQ, Paschon DE, Miranda E, Ordóñez A, Hannan NR, Rouhani FJ, Darche S, Alexander G, Marciniak SJ, Fusaki N, Hasegawa M, Holmes MC, Di Santo JP, Lomas DA, Bradley A, Vallier L (October 2011). "Targeted gene correction of α1-antitrypsin deficiency in induced pluripotent stem cells". Nature. 478 (7369): 391–394. Bibcode:2011Natur.478..391Y. doi:10.1038/nature10424. PMC 3198846. PMID 21993621.
- ^ Mallya M, Phillips RL, Saldanha SA, Gooptu B, Brown SC, Termine DJ, Shirvani AM, Wu Y, Sifers RN, Abagyan R, Lomas DA (November 2007). "Small molecules block the polymerization of Z alpha1-antitrypsin and increase the clearance of intracellular aggregates". Journal of Medicinal Chemistry. 50 (22): 5357–5363. doi:10.1021/jm070687z. PMC 2631427. PMID 17918823.
- ^ Gosai SJ, Kwak JH, Luke CJ, Long OS, King DE, Kovatch KJ, Johnston PA, Shun TY, Lazo JS, Perlmutter DH, Silverman GA, Pak SC (November 2010). "Automated high-content live animal drug screening using C. elegans expressing the aggregation prone serpin α1-antitrypsin Z". PLOS ONE. 5 (11): e15460. Bibcode:2010PLoSO...515460G. doi:10.1371/journal.pone.0015460. PMC 2980495. PMID 21103396.
- ^ Cabrita LD, Irving JA, Pearce MC, Whisstock JC, Bottomley SP (September 2007). "Aeropin from the extremophile Pyrobaculum aerophilum bypasses the serpin misfolding trap". The Journal of Biological Chemistry. 282 (37): 26802–26809. doi:10.1074/jbc.M705020200. PMID 17635906.
- ^ Fluhr R, Lampl N, Roberts TH (May 2012). "Serpin protease inhibitors in plant biology". Physiologia Plantarum. 145 (1): 95–102. doi:10.1111/j.1399-3054.2011.01540.x. PMID 22085334.
- ^ Stoller JK, Aboussouan LS (2005). "Alpha1-antitrypsin deficiency". Lancet. 365 (9478): 2225–2236. doi:10.1016/S0140-6736(05)66781-5. PMID 15978931. S2CID 54415934.
- ^ Münch J, Ständker L, Adermann K, Schulz A, Schindler M, Chinnadurai R, Pöhlmann S, Chaipan C, Biet T, Peters T, Meyer B, Wilhelm D, Lu H, Jing W, Jiang S, Forssmann WG, Kirchhoff F (April 2007). "Discovery and optimization of a natural HIV-1 entry inhibitor targeting the gp41 fusion peptide". Cell. 129 (2): 263–275. doi:10.1016/j.cell.2007.02.042. PMID 17448989.
- ^ Gooptu B, Dickens JA, Lomas DA (February 2014). "The molecular and cellular pathology of α₁-antitrypsin deficiency". Trends in Molecular Medicine. 20 (2): 116–127. doi:10.1016/j.molmed.2013.10.007. PMID 24374162.
- ^ Seixas S, Suriano G, Carvalho F, Seruca R, Rocha J, Di Rienzo A (February 2007). "Sequence diversity at the proximal 14q32.1 SERPIN subcluster: evidence for natural selection favoring the pseudogenization of SERPINA2". Molecular Biology and Evolution. 24 (2): 587–598. doi:10.1093/molbev/msl187. PMID 17135331.
- ^ Kalsheker NA (September 1996). "Alpha 1-antichymotrypsin". The International Journal of Biochemistry & Cell Biology. 28 (9): 961–964. doi:10.1016/1357-2725(96)00032-5. PMID 8930118. S2CID 11230631.
- ^ Santamaria M, Pardo-Saganta A, Alvarez-Asiain L, Di Scala M, Qian C, Prieto J, Avila MA (April 2013). "Nuclear α1-antichymotrypsin promotes chromatin condensation and inhibits proliferation of human hepatocellular carcinoma cells". Gastroenterology. 144 (4): 818–828.e4. doi:10.1053/j.gastro.2012.12.029. PMID 23295442.
- ^ Zhang S, Janciauskiene S (April 2002). "Multi-functional capability of proteins: alpha1-antichymotrypsin and the correlation with Alzheimer's disease". Journal of Alzheimer's Disease. 4 (2): 115–122. doi:10.3233/JAD-2002-4206. PMID 12214135.
- ^ Chao J, Stallone JN, Liang YM, Chen LM, Wang DZ, Chao L (July 1997). "Kallistatin is a potent new vasodilator". The Journal of Clinical Investigation. 100 (1): 11–17. doi:10.1172/JCI119502. PMC 508159. PMID 9202051.
- ^ Miao RQ, Agata J, Chao L, Chao J (November 2002). "Kallistatin is a new inhibitor of angiogenesis and tumor growth". Blood. 100 (9): 3245–3252. doi:10.1182/blood-2002-01-0185. PMID 12384424.
- ^ Liu Y, Bledsoe G, Hagiwara M, Shen B, Chao L, Chao J (October 2012). "Depletion of endogenous kallistatin exacerbates renal and cardiovascular oxidative stress, inflammation, and organ remodeling". American Journal of Physiology. Renal Physiology. 303 (8): F1230–F1238. doi:10.1152/ajprenal.00257.2012. PMC 3469672. PMID 22811485.
- ^ Geiger M (March 2007). "Protein C inhibitor, a serpin with functions in- and outside vascular biology". Thrombosis and Haemostasis. 97 (3): 343–347. doi:10.1160/th06-09-0488. PMID 17334499. S2CID 4828322.
- ^ Baumgärtner P, Geiger M, Zieseniss S, Malleier J, Huntington JA, Hochrainer K, Bielek E, Stoeckelhuber M, Lauber K, Scherfeld D, Schwille P, Wäldele K, Beyer K, Engelmann B (November 2007). "Phosphatidylethanolamine critically supports internalization of cell-penetrating protein C inhibitor". The Journal of Cell Biology. 179 (4): 793–804. doi:10.1083/jcb.200707165. PMC 2080921. PMID 18025309.
- ^ Uhrin P, Dewerchin M, Hilpert M, Chrenek P, Schöfer C, Zechmeister-Machhart M, Krönke G, Vales A, Carmeliet P, Binder BR, Geiger M (December 2000). "Disruption of the protein C inhibitor gene results in impaired spermatogenesis and male infertility". The Journal of Clinical Investigation. 106 (12): 1531–1539. doi:10.1172/JCI10768. PMC 381472. PMID 11120760.
- ^ Han MH, Hwang SI, Roy DB, Lundgren DH, Price JV, Ousman SS, Fernald GH, Gerlitz B, Robinson WH, Baranzini SE, Grinnell BW, Raine CS, Sobel RA, Han DK, Steinman L (February 2008). "Proteomic analysis of active multiple sclerosis lesions reveals therapeutic targets". Nature. 451 (7182): 1076–1081. Bibcode:2008Natur.451.1076H. doi:10.1038/nature06559. PMID 18278032. S2CID 4421395.
- ^ Torpy DJ, Ho JT (August 2007). "Corticosteroid-binding globulin gene polymorphisms: clinical implications and links to idiopathic chronic fatigue disorders". Clinical Endocrinology. 67 (2): 161–167. doi:10.1111/j.1365-2265.2007.02890.x. PMID 17547679. S2CID 43352358.
- ^ Bartalena L, Robbins J (1992). "Variations in thyroid hormone transport proteins and their clinical implications". Thyroid. 2 (3): 237–245. doi:10.1089/thy.1992.2.237. PMID 1422238.
- ^ Persani L (September 2012). "Clinical review: Central hypothyroidism: pathogenic, diagnostic, and therapeutic challenges". The Journal of Clinical Endocrinology and Metabolism. 97 (9): 3068–3078. doi:10.1210/jc.2012-1616. PMID 22851492.
- ^ Kumar R, Singh VP, Baker KM (July 2007). "The intracellular renin-angiotensin system: a new paradigm". Trends in Endocrinology and Metabolism. 18 (5): 208–214. doi:10.1016/j.tem.2007.05.001. PMID 17509892. S2CID 24041932.
- ^ Tanimoto K, Sugiyama F, Goto Y, Ishida J, Takimoto E, Yagami K, Fukamizu A, Murakami K (December 1994). "Angiotensinogen-deficient mice with hypotension". The Journal of Biological Chemistry. 269 (50): 31334–31337. doi:10.1016/S0021-9258(18)31697-1. PMID 7989296.
- ^ Jeunemaitre X, Gimenez-Roqueplo AP, Célérier J, Corvol P (1999). "Angiotensinogen variants and human hypertension". Current Hypertension Reports. 1 (1): 31–41. doi:10.1007/s11906-999-0071-0. PMID 10981040. S2CID 42614761.
- ^ Sethi AA, Nordestgaard BG, Tybjaerg-Hansen A (July 2003). "Angiotensinogen gene polymorphism, plasma angiotensinogen, and risk of hypertension and ischemic heart disease: a meta-analysis". Arteriosclerosis, Thrombosis, and Vascular Biology. 23 (7): 1269–1275. doi:10.1161/01.ATV.0000079007.40884.5C. PMID 12805070.
- ^ Dickson ME, Sigmund CD (July 2006). "Genetic basis of hypertension: revisiting angiotensinogen". Hypertension. 48 (1): 14–20. doi:10.1161/01.HYP.0000227932.13687.60. PMID 16754793.
- ^ Frazer JK, Jackson DG, Gaillard JP, Lutter M, Liu YJ, Banchereau J, Capra JD, Pascual V (October 2000). "Identification of centerin: a novel human germinal center B cell-restricted serpin". European Journal of Immunology. 30 (10): 3039–3048. doi:10.1002/1521-4141(200010)30:10<3039::AID-IMMU3039>3.0.CO;2-H. PMID 11069088.
- ^ Paterson MA, Horvath AJ, Pike RN, Coughlin PB (August 2007). "Molecular characterization of centerin, a germinal centre cell serpin". The Biochemical Journal. 405 (3): 489–494. doi:10.1042/BJ20070174. PMC 2267310. PMID 17447896.
- ^ Paterson MA, Hosking PS, Coughlin PB (July 2008). "Expression of the serpin centerin defines a germinal center phenotype in B-cell lymphomas". American Journal of Clinical Pathology. 130 (1): 117–126. doi:10.1309/9QKE68QU7B825A3U. PMID 18550480.
- ^ Ashton-Rickardt PG (April 2013). "An emerging role for Serine Protease Inhibitors in T lymphocyte immunity and beyond". Immunology Letters. 152 (1): 65–76. doi:10.1016/j.imlet.2013.04.004. PMID 23624075.
- ^ Han X, Fiehler R, Broze GJ (November 2000). "Characterization of the protein Z-dependent protease inhibitor". Blood. 96 (9): 3049–3055. doi:10.1182/blood.V96.9.3049. PMID 11049983.
- ^ Hida K, Wada J, Eguchi J, Zhang H, Baba M, Seida A, Hashimoto I, Okada T, Yasuhara A, Nakatsuka A, Shikata K, Hourai S, Futami J, Watanabe E, Matsuki Y, Hiramatsu R, Akagi S, Makino H, Kanwar YS (July 2005). "Visceral adipose tissue-derived serine protease inhibitor: a unique insulin-sensitizing adipocytokine in obesity". Proceedings of the National Academy of Sciences of the United States of America. 102 (30): 10610–10615. Bibcode:2005PNAS..10210610H. doi:10.1073/pnas.0504703102. PMC 1180799. PMID 16030142.
- ^ Feng R, Li Y, Wang C, Luo C, Liu L, Chuo F, Li Q, Sun C (October 2014). "Higher vaspin levels in subjects with obesity and type 2 diabetes mellitus: a meta-analysis". Diabetes Research and Clinical Practice. 106 (1): 88–94. doi:10.1016/j.diabres.2014.07.026. PMID 25151227.
- ^ Remold-O'Donnell E, Chin J, Alberts M (June 1992). "Sequence and molecular characterization of human monocyte/neutrophil elastase inhibitor". Proceedings of the National Academy of Sciences of the United States of America. 89 (12): 5635–5639. Bibcode:1992PNAS...89.5635R. doi:10.1073/pnas.89.12.5635. PMC 49347. PMID 1376927.
- ^ Benarafa C, Priebe GP, Remold-O'Donnell E (August 2007). "The neutrophil serine protease inhibitor serpinb1 preserves lung defense functions in Pseudomonas aeruginosa infection". The Journal of Experimental Medicine. 204 (8): 1901–1909. doi:10.1084/jem.20070494. PMC 2118684. PMID 17664292.
- ^ Antalis TM, La Linn M, Donnan K, Mateo L, Gardner J, Dickinson JL, Buttigieg K, Suhrbier A (June 1998). "The serine proteinase inhibitor (serpin) plasminogen activation inhibitor type 2 protects against viral cytopathic effects by constitutive interferon alpha/beta priming". The Journal of Experimental Medicine. 187 (11): 1799–1811. doi:10.1084/jem.187.11.1799. PMC 2212304. PMID 9607921.
- ^ Zhao A, Yang Z, Sun R, Grinchuk V, Netzel-Arnett S, Anglin IE, Driesbaugh KH, Notari L, Bohl JA, Madden KB, Urban JF, Antalis TM, Shea-Donohue T (June 2013). "SerpinB2 is critical to Th2 immunity against enteric nematode infection". Journal of Immunology. 190 (11): 5779–5787. doi:10.4049/jimmunol.1200293. PMC 4068334. PMID 23630350.
- ^ Dougherty KM, Pearson JM, Yang AY, Westrick RJ, Baker MS, Ginsburg D (January 1999). "The plasminogen activator inhibitor-2 gene is not required for normal murine development or survival". Proceedings of the National Academy of Sciences of the United States of America. 96 (2): 686–691. Bibcode:1999PNAS...96..686D. doi:10.1073/pnas.96.2.686. PMC 15197. PMID 9892694.
- ^ Takeda A, Yamamoto T, Nakamura Y, Takahashi T, Hibino T (February 1995). "Squamous cell carcinoma antigen is a potent inhibitor of cysteine proteinase cathepsin L". FEBS Letters. 359 (1): 78–80. Bibcode:1995FEBSL.359...78T. doi:10.1016/0014-5793(94)01456-b. PMID 7851535. S2CID 35146299.
- ^ a b Turato C, Pontisso P (March 2015). "SERPINB3 (serpin peptidase inhibitor, clade B (ovalbumin), member 3)". Atlas of Genetics and Cytogenetics in Oncology and Haematology. 19 (3): 202–209. doi:10.4267/2042/56413 (inactive 1 November 2024). PMC 4430857. PMID 25984243.
{{cite journal}}: CS1 maint: DOI inactive as of November 2024 (link) - ^ a b Sivaprasad U, Askew DJ, Ericksen MB, Gibson AM, Stier MT, Brandt EB, Bass SA, Daines MO, Chakir J, Stringer KF, Wert SE, Whitsett JA, Le Cras TD, Wills-Karp M, Silverman GA, Khurana Hershey GK (January 2011). "A nonredundant role for mouse Serpinb3a in the induction of mucus production in asthma". The Journal of Allergy and Clinical Immunology. 127 (1): 254–61, 261.e1–6. doi:10.1016/j.jaci.2010.10.009. PMC 3058372. PMID 21126757.
- ^ Schick C, Kamachi Y, Bartuski AJ, Cataltepe S, Schechter NM, Pemberton PA, Silverman GA (January 1997). "Squamous cell carcinoma antigen 2 is a novel serpin that inhibits the chymotrypsin-like proteinases cathepsin G and mast cell chymase". The Journal of Biological Chemistry. 272 (3): 1849–1855. doi:10.1074/jbc.272.3.1849. PMID 8999871.
- ^ Teoh SS, Whisstock JC, Bird PI (April 2010). "Maspin (SERPINB5) is an obligate intracellular serpin". The Journal of Biological Chemistry. 285 (14): 10862–10869. doi:10.1074/jbc.M109.073171. PMC 2856292. PMID 20123984.
- ^ Zou Z, Anisowicz A, Hendrix MJ, Thor A, Neveu M, Sheng S, Rafidi K, Seftor E, Sager R (January 1994). "Maspin, a serpin with tumor-suppressing activity in human mammary epithelial cells". Science. 263 (5146): 526–529. Bibcode:1994Sci...263..526Z. doi:10.1126/science.8290962. PMID 8290962.
- ^ a b c Teoh SS, Vieusseux J, Prakash M, Berkowicz S, Luu J, Bird CH, Law RH, Rosado C, Price JT, Whisstock JC, Bird PI (2014). "Maspin is not required for embryonic development or tumour suppression". Nature Communications. 5: 3164. Bibcode:2014NatCo...5.3164T. doi:10.1038/ncomms4164. PMC 3905777. PMID 24445777.
- ^ Gao F, Shi HY, Daughty C, Cella N, Zhang M (April 2004). "Maspin plays an essential role in early embryonic development". Development. 131 (7): 1479–1489. doi:10.1242/dev.01048. PMID 14985257.
- ^ Scott FL, Hirst CE, Sun J, Bird CH, Bottomley SP, Bird PI (March 1999). "The intracellular serpin proteinase inhibitor 6 is expressed in monocytes and granulocytes and is a potent inhibitor of the azurophilic granule protease, cathepsin G". Blood. 93 (6): 2089–2097. doi:10.1182/blood.V93.6.2089.406k10_2089_2097. PMID 10068683.
- ^ Tan J, Prakash MD, Kaiserman D, Bird PI (July 2013). "Absence of SERPINB6A causes sensorineural hearing loss with multiple histopathologies in the mouse inner ear". The American Journal of Pathology. 183 (1): 49–59. doi:10.1016/j.ajpath.2013.03.009. PMID 23669344.
- ^ Scarff KL, Ung KS, Nandurkar H, Crack PJ, Bird CH, Bird PI (May 2004). "Targeted disruption of SPI3/Serpinb6 does not result in developmental or growth defects, leukocyte dysfunction, or susceptibility to stroke". Molecular and Cellular Biology. 24 (9): 4075–4082. doi:10.1128/MCB.24.9.4075-4082.2004. PMC 387772. PMID 15082799.
- ^ Sirmaci A, Erbek S, Price J, Huang M, Duman D, Cengiz FB, Bademci G, Tokgöz-Yilmaz S, Hişmi B, Ozdağ H, Oztürk B, Kulaksizoğlu S, Yildirim E, Kokotas H, Grigoriadou M, Petersen MB, Shahin H, Kanaan M, King MC, Chen ZY, Blanton SH, Liu XZ, Zuchner S, Akar N, Tekin M (May 2010). "A truncating mutation in SERPINB6 is associated with autosomal-recessive nonsyndromic sensorineural hearing loss". American Journal of Human Genetics. 86 (5): 797–804. doi:10.1016/j.ajhg.2010.04.004. PMC 2869020. PMID 20451170.
- ^ Miyata T, Inagi R, Nangaku M, Imasawa T, Sato M, Izuhara Y, Suzuki D, Yoshino A, Onogi H, Kimura M, Sugiyama S, Kurokawa K (March 2002). "Overexpression of the serpin megsin induces progressive mesangial cell proliferation and expansion". The Journal of Clinical Investigation. 109 (5): 585–593. doi:10.1172/JCI14336. PMC 150894. PMID 11877466.
- ^ a b Miyata T, Li M, Yu X, Hirayama N (May 2007). "Megsin gene: its genomic analysis, pathobiological functions, and therapeutic perspectives". Current Genomics. 8 (3): 203–208. doi:10.2174/138920207780833856. PMC 2435355. PMID 18645605.
- ^ Kubo A (August 2014). "Nagashima-type palmoplantar keratosis: a common Asian type caused by SERPINB7 protease inhibitor deficiency". The Journal of Investigative Dermatology. 134 (8): 2076–2079. doi:10.1038/jid.2014.156. PMID 25029323.
- ^ Dahlen JR, Jean F, Thomas G, Foster DC, Kisiel W (January 1998). "Inhibition of soluble recombinant furin by human proteinase inhibitor 8". The Journal of Biological Chemistry. 273 (4): 1851–1854. doi:10.1074/jbc.273.4.1851. PMID 9442015.
- ^ Sun J, Bird CH, Sutton V, McDonald L, Coughlin PB, De Jong TA, Trapani JA, Bird PI (November 1996). "A cytosolic granzyme B inhibitor related to the viral apoptotic regulator cytokine response modifier A is present in cytotoxic lymphocytes". The Journal of Biological Chemistry. 271 (44): 27802–27809. doi:10.1074/jbc.271.44.27802. PMID 8910377.
- ^ Zhang M, Park SM, Wang Y, Shah R, Liu N, Murmann AE, Wang CR, Peter ME, Ashton-Rickardt PG (April 2006). "Serine protease inhibitor 6 protects cytotoxic T cells from self-inflicted injury by ensuring the integrity of cytotoxic granules". Immunity. 24 (4): 451–461. doi:10.1016/j.immuni.2006.02.002. PMID 16618603.
- ^ Rizzitelli A, Meuter S, Vega Ramos J, Bird CH, Mintern JD, Mangan MS, Villadangos J, Bird PI (October 2012). "Serpinb9 (Spi6)-deficient mice are impaired in dendritic cell-mediated antigen cross-presentation". Immunology and Cell Biology. 90 (9): 841–851. doi:10.1038/icb.2012.29. PMID 22801574. S2CID 39276036.
- ^ Riewald M, Chuang T, Neubauer A, Riess H, Schleef RR (February 1998). "Expression of bomapin, a novel human serpin, in normal/malignant hematopoiesis and in the monocytic cell lines THP-1 and AML-193". Blood. 91 (4): 1256–1262. doi:10.1182/blood.V91.4.1256. PMID 9454755.
- ^ a b Askew DJ, Cataltepe S, Kumar V, Edwards C, Pace SM, Howarth RN, Pak SC, Askew YS, Brömme D, Luke CJ, Whisstock JC, Silverman GA (August 2007). "SERPINB11 is a new noninhibitory intracellular serpin. Common single nucleotide polymorphisms in the scaffold impair conformational change". The Journal of Biological Chemistry. 282 (34): 24948–24960. doi:10.1074/jbc.M703182200. PMID 17562709.
- ^ Finno CJ, Stevens C, Young A, Affolter V, Joshi NA, Ramsay S, Bannasch DL (April 2015). "SERPINB11 frameshift variant associated with novel hoof specific phenotype in Connemara ponies". PLOS Genetics. 11 (4): e1005122. doi:10.1371/journal.pgen.1005122. PMC 4395385. PMID 25875171.
- ^ Askew YS, Pak SC, Luke CJ, Askew DJ, Cataltepe S, Mills DR, Kato H, Lehoczky J, Dewar K, Birren B, Silverman GA (December 2001). "SERPINB12 is a novel member of the human ov-serpin family that is widely expressed and inhibits trypsin-like serine proteinases". The Journal of Biological Chemistry. 276 (52): 49320–49330. doi:10.1074/jbc.M108879200. PMID 11604408.
- ^ Welss T, Sun J, Irving JA, Blum R, Smith AI, Whisstock JC, Pike RN, von Mikecz A, Ruzicka T, Bird PI, Abts HF (June 2003). "Hurpin is a selective inhibitor of lysosomal cathepsin L and protects keratinocytes from ultraviolet-induced apoptosis". Biochemistry. 42 (24): 7381–7389. doi:10.1021/bi027307q. PMID 12809493.
- ^ Ishiguro K, Kojima T, Kadomatsu K, Nakayama Y, Takagi A, Suzuki M, Takeda N, Ito M, Yamamoto K, Matsushita T, Kusugami K, Muramatsu T, Saito H (October 2000). "Complete antithrombin deficiency in mice results in embryonic lethality". The Journal of Clinical Investigation. 106 (7): 873–878. doi:10.1172/JCI10489. PMC 517819. PMID 11018075.
- ^ Patnaik MM, Moll S (November 2008). "Inherited antithrombin deficiency: a review". Haemophilia. 14 (6): 1229–1239. doi:10.1111/j.1365-2516.2008.01830.x. PMID 19141163. S2CID 20768425.
- ^ Huntington JA (July 2011). "Serpin structure, function and dysfunction". Journal of Thrombosis and Haemostasis. 9 (Suppl 1): 26–34. doi:10.1111/j.1538-7836.2011.04360.x. PMID 21781239. S2CID 1020630.
- ^ Vicente CP, He L, Pavão MS, Tollefsen DM (December 2004). "Antithrombotic activity of dermatan sulfate in heparin cofactor II-deficient mice". Blood. 104 (13): 3965–3970. doi:10.1182/blood-2004-02-0598. PMID 15315969.
- ^ Aihara K, Azuma H, Akaike M, Ikeda Y, Sata M, Takamori N, Yagi S, Iwase T, Sumitomo Y, Kawano H, Yamada T, Fukuda T, Matsumoto T, Sekine K, Sato T, Nakamichi Y, Yamamoto Y, Yoshimura K, Watanabe T, Nakamura T, Oomizu A, Tsukada M, Hayashi H, Sudo T, Kato S, Matsumoto T (June 2007). "Strain-dependent embryonic lethality and exaggerated vascular remodeling in heparin cofactor II-deficient mice". The Journal of Clinical Investigation. 117 (6): 1514–1526. doi:10.1172/JCI27095. PMC 1878511. PMID 17549254.
- ^ Cale JM, Lawrence DA (September 2007). "Structure-function relationships of plasminogen activator inhibitor-1 and its potential as a therapeutic agent". Current Drug Targets. 8 (9): 971–981. doi:10.2174/138945007781662337. PMID 17896949.
- ^ Lino MM, Atanasoski S, Kvajo M, Fayard B, Moreno E, Brenner HR, Suter U, Monard D (April 2007). "Mice lacking protease nexin-1 show delayed structural and functional recovery after sciatic nerve crush". The Journal of Neuroscience. 27 (14): 3677–3685. doi:10.1523/JNEUROSCI.0277-07.2007. PMC 6672422. PMID 17409231.
- ^ Murer V, Spetz JF, Hengst U, Altrogge LM, de Agostini A, Monard D (March 2001). "Male fertility defects in mice lacking the serine protease inhibitor protease nexin-1". Proceedings of the National Academy of Sciences of the United States of America. 98 (6): 3029–3033. Bibcode:2001PNAS...98.3029M. doi:10.1073/pnas.051630698. PMC 30601. PMID 11248026.
- ^ Lüthi A, Van der Putten H, Botteri FM, Mansuy IM, Meins M, Frey U, Sansig G, Portet C, Schmutz M, Schröder M, Nitsch C, Laurent JP, Monard D (June 1997). "Endogenous serine protease inhibitor modulates epileptic activity and hippocampal long-term potentiation". The Journal of Neuroscience. 17 (12): 4688–4699. doi:10.1523/JNEUROSCI.17-12-04688.1997. PMC 6573330. PMID 9169529.
- ^ a b Doll JA, Stellmach VM, Bouck NP, Bergh AR, Lee C, Abramson LP, Cornwell ML, Pins MR, Borensztajn J, Crawford SE (June 2003). "Pigment epithelium-derived factor regulates the vasculature and mass of the prostate and pancreas". Nature Medicine. 9 (6): 774–780. doi:10.1038/nm870. PMID 12740569. S2CID 5967666.
- ^ Becerra SP, Perez-Mediavilla LA, Weldon JE, Locatelli-Hoops S, Senanayake P, Notari L, Notario V, Hollyfield JG (November 2008). "Pigment epithelium-derived factor binds to hyaluronan. Mapping of a hyaluronan binding site". The Journal of Biological Chemistry. 283 (48): 33310–33320. doi:10.1074/jbc.M801287200. PMC 2586245. PMID 18805795.
- ^ Andreu-Agulló C, Morante-Redolat JM, Delgado AC, Fariñas I (December 2009). "Vascular niche factor PEDF modulates Notch-dependent stemness in the adult subependymal zone". Nature Neuroscience. 12 (12): 1514–1523. doi:10.1038/nn.2437. PMID 19898467. S2CID 5332822.
- ^ Wiman B, Collen D (September 1979). "On the mechanism of the reaction between human alpha 2-antiplasmin and plasmin". The Journal of Biological Chemistry. 254 (18): 9291–9297. doi:10.1016/S0021-9258(19)86843-6. PMID 158022.
- ^ Lijnen HR, Okada K, Matsuo O, Collen D, Dewerchin M (April 1999). "Alpha2-antiplasmin gene deficiency in mice is associated with enhanced fibrinolytic potential without overt bleeding". Blood. 93 (7): 2274–2281. doi:10.1182/blood.V93.7.2274. PMID 10090937.
- ^ Carpenter SL, Mathew P (November 2008). "Alpha2-antiplasmin and its deficiency: fibrinolysis out of balance". Haemophilia. 14 (6): 1250–1254. doi:10.1111/j.1365-2516.2008.01766.x. PMID 19141165. S2CID 205295156.
- ^ Favier R, Aoki N, de Moerloose P (July 2001). "Congenital alpha(2)-plasmin inhibitor deficiencies: a review". British Journal of Haematology. 114 (1): 4–10. doi:10.1046/j.1365-2141.2001.02845.x. PMID 11472338. S2CID 71010865.
- ^ Beinrohr L, Harmat V, Dobó J, Lörincz Z, Gál P, Závodszky P (July 2007). "C1 inhibitor serpin domain structure reveals the likely mechanism of heparin potentiation and conformational disease". The Journal of Biological Chemistry. 282 (29): 21100–21109. doi:10.1074/jbc.M700841200. hdl:10831/92835. PMID 17488724.
- ^ Mollnes TE, Jokiranta TS, Truedsson L, Nilsson B, Rodriguez de Cordoba S, Kirschfink M (September 2007). "Complement analysis in the 21st century". Molecular Immunology. 44 (16): 3838–3849. doi:10.1016/j.molimm.2007.06.150. hdl:10261/61732. PMID 17768101.
- ^ Triggianese P, Chimenti MS, Toubi E, Ballanti E, Guarino MD, Perricone C, Perricone R (August 2015). "The autoimmune side of hereditary angioedema: insights on the pathogenesis". Autoimmunity Reviews. 14 (8): 665–669. doi:10.1016/j.autrev.2015.03.006. PMID 25827463.
- ^ Nagai N, Hosokawa M, Itohara S, Adachi E, Matsushita T, Hosokawa N, Nagata K (September 2000). "Embryonic lethality of molecular chaperone hsp47 knockout mice is associated with defects in collagen biosynthesis". The Journal of Cell Biology. 150 (6): 1499–1506. doi:10.1083/jcb.150.6.1499. PMC 2150697. PMID 10995453.
- ^ Marini JC, Reich A, Smith SM (August 2014). "Osteogenesis imperfecta due to mutations in non-collagenous genes: lessons in the biology of bone formation". Current Opinion in Pediatrics. 26 (4): 500–507. doi:10.1097/MOP.0000000000000117. PMC 4183132. PMID 25007323.
- ^ Byers PH, Pyott SM (1 January 2012). "Recessively inherited forms of osteogenesis imperfecta". Annual Review of Genetics. 46: 475–497. doi:10.1146/annurev-genet-110711-155608. PMID 23145505.
- ^ Osterwalder T, Cinelli P, Baici A, Pennella A, Krueger SR, Schrimpf SP, Meins M, Sonderegger P (January 1998). "The axonally secreted serine proteinase inhibitor, neuroserpin, inhibits plasminogen activators and plasmin but not thrombin". The Journal of Biological Chemistry. 273 (4): 2312–2321. doi:10.1074/jbc.273.4.2312. PMID 9442076.
- ^ Crowther DC (July 2002). "Familial conformational diseases and dementias". Human Mutation. 20 (1): 1–14. doi:10.1002/humu.10100. PMID 12112652. S2CID 22326349.
- ^ Belorgey D, Hägglöf P, Karlsson-Li S, Lomas DA (1 March 2007). "Protein misfolding and the serpinopathies". Prion. 1 (1): 15–20. doi:10.4161/pri.1.1.3974. PMC 2633702. PMID 19164889.
- ^ Ozaki K, Nagata M, Suzuki M, Fujiwara T, Miyoshi Y, Ishikawa O, Ohigashi H, Imaoka S, Takahashi E, Nakamura Y (July 1998). "Isolation and characterization of a novel human pancreas-specific gene, pancpin, that is down-regulated in pancreatic cancer cells". Genes, Chromosomes & Cancer. 22 (3): 179–185. doi:10.1002/(SICI)1098-2264(199807)22:3<179::AID-GCC3>3.0.CO;2-T. PMID 9624529. S2CID 9525421.
- ^ Loftus SK, Cannons JL, Incao A, Pak E, Chen A, Zerfas PM, Bryant MA, Biesecker LG, Schwartzberg PL, Pavan WJ (September 2005). "Acinar cell apoptosis in Serpini2-deficient mice models pancreatic insufficiency". PLOS Genetics. 1 (3): e38. doi:10.1371/journal.pgen.0010038. PMC 1231717. PMID 16184191.
- ^ Padua MB, Kowalski AA, Cañas MY, Hansen PJ (February 2010). "The molecular phylogeny of uterine serpins and its relationship to evolution of placentation". FASEB Journal. 24 (2): 526–537. doi:10.1096/fj.09-138453. PMID 19825977. S2CID 9248169.
- ^ Padua MB, Hansen PJ (October 2010). "Evolution and function of the uterine serpins (SERPINA14)". American Journal of Reproductive Immunology. 64 (4): 265–274. doi:10.1111/j.1600-0897.2010.00901.x. PMID 20678169.
- ^ a b Reichhart JM (December 2005). "Tip of another iceberg: Drosophila serpins". Trends in Cell Biology. 15 (12): 659–665. doi:10.1016/j.tcb.2005.10.001. PMID 16260136.
- ^ Tang H, Kambris Z, Lemaitre B, Hashimoto C (October 2008). "A serpin that regulates immune melanization in the respiratory system of Drosophila". Developmental Cell. 15 (4): 617–626. doi:10.1016/j.devcel.2008.08.017. PMC 2671232. PMID 18854145.
- ^ Scherfer C, Tang H, Kambris Z, Lhocine N, Hashimoto C, Lemaitre B (November 2008). "Drosophila Serpin-28D regulates hemolymph phenoloxidase activity and adult pigmentation". Developmental Biology. 323 (2): 189–196. doi:10.1016/j.ydbio.2008.08.030. PMID 18801354.
- ^ Rushlow C (January 2004). "Dorsoventral patterning: a serpin pinned down at last". Current Biology. 14 (1): R16–R18. Bibcode:2004CBio...14..R16R. doi:10.1016/j.cub.2003.12.015. PMID 14711428.
- ^ Ligoxygakis P, Roth S, Reichhart JM (December 2003). "A serpin regulates dorsal-ventral axis formation in the Drosophila embryo". Current Biology. 13 (23): 2097–2102. Bibcode:2003CBio...13.2097L. doi:10.1016/j.cub.2003.10.062. PMID 14654000.
- ^ Jiang R, Zhang B, Kurokawa K, So YI, Kim EH, Hwang HO, Lee JH, Shiratsuchi A, Zhang J, Nakanishi Y, Lee HS, Lee BL (October 2011). "93-kDa twin-domain serine protease inhibitor (Serpin) has a regulatory function on the beetle Toll proteolytic signaling cascade". The Journal of Biological Chemistry. 286 (40): 35087–35095. doi:10.1074/jbc.M111.277343. PMC 3186399. PMID 21862574.
- ^ a b Pak SC, Kumar V, Tsu C, Luke CJ, Askew YS, Askew DJ, Mills DR, Brömme D, Silverman GA (April 2004). "SRP-2 is a cross-class inhibitor that participates in postembryonic development of the nematode Caenorhabditis elegans: initial characterization of the clade L serpins". The Journal of Biological Chemistry. 279 (15): 15448–15459. doi:10.1074/jbc.M400261200. PMID 14739286.
- ^ Luke CJ, Pak SC, Askew YS, Naviglia TL, Askew DJ, Nobar SM, Vetica AC, Long OS, Watkins SC, Stolz DB, Barstead RJ, Moulder GL, Brömme D, Silverman GA (September 2007). "An intracellular serpin regulates necrosis by inhibiting the induction and sequelae of lysosomal injury". Cell. 130 (6): 1108–1119. doi:10.1016/j.cell.2007.07.013. PMC 2128786. PMID 17889653.
- ^ Hejgaard J, Rasmussen SK, Brandt A, SvendsenI (1985). "Sequence homology between barley endosperm protein Z and protease inhibitors of the alpha-1-antitrypsin family". FEBS Lett. 180 (1): 89–94. Bibcode:1985FEBSL.180...89H. doi:10.1016/0014-5793(85)80238-6. S2CID 84790014.
- ^ Dahl SW, Rasmussen SK, Petersen LC, Hejgaard J (September 1996). "Inhibition of coagulation factors by recombinant barley serpin BSZx". FEBS Letters. 394 (2): 165–168. Bibcode:1996FEBSL.394..165D. doi:10.1016/0014-5793(96)00940-4. PMID 8843156.
- ^ Hejgaard J (January 2001). "Inhibitory serpins from rye grain with glutamine as P1 and P2 residues in the reactive center". FEBS Letters. 488 (3): 149–153. Bibcode:2001FEBSL.488..149H. doi:10.1016/S0014-5793(00)02425-X. PMID 11163762. S2CID 27933086.
- ^ Ostergaard H, Rasmussen SK, Roberts TH, Hejgaard J (October 2000). "Inhibitory serpins from wheat grain with reactive centers resembling glutamine-rich repeats of prolamin storage proteins. Cloning and characterization of five major molecular forms". The Journal of Biological Chemistry. 275 (43): 33272–33279. doi:10.1074/jbc.M004633200. PMID 10874043.
- ^ a b Yoo BC, Aoki K, Xiang Y, Campbell LR, Hull RJ, Xoconostle-Cázares B, Monzer J, Lee JY, Ullman DE, Lucas WJ (November 2000). "Characterization of cucurbita maxima phloem serpin-1 (CmPS-1). A developmentally regulated elastase inhibitor". The Journal of Biological Chemistry. 275 (45): 35122–35128. doi:10.1074/jbc.M006060200. PMID 10960478.
- ^ la Cour Petersen M, Hejgaard J, Thompson GA, Schulz A (December 2005). "Cucurbit phloem serpins are graft-transmissible and appear to be resistant to turnover in the sieve element-companion cell complex". Journal of Experimental Botany. 56 (422): 3111–3120. doi:10.1093/jxb/eri308. PMID 16246856.
- ^ Roberts TH, Hejgaard J (February 2008). "Serpins in plants and green algae". Functional & Integrative Genomics. 8 (1): 1–27. doi:10.1007/s10142-007-0059-2. PMID 18060440. S2CID 22960858.
- ^ Lampl N, Alkan N, Davydov O, Fluhr R (May 2013). "Set-point control of RD21 protease activity by AtSerpin1 controls cell death in Arabidopsis". The Plant Journal. 74 (3): 498–510. doi:10.1111/tpj.12141. PMID 23398119.
- ^ Ahn JW, Atwell BJ, Roberts TH (May 2009). "Serpin genes AtSRP2 and AtSRP3 are required for normal growth sensitivity to a DNA alkylating agent in Arabidopsis". BMC Plant Biology. 9: 52. doi:10.1186/1471-2229-9-52. PMC 2689219. PMID 19426562.
- ^ a b Kang S, Barak Y, Lamed R, Bayer EA, Morrison M (June 2006). "The functional repertoire of prokaryote cellulosomes includes the serpin superfamily of serine proteinase inhibitors". Molecular Microbiology. 60 (6): 1344–1354. doi:10.1111/j.1365-2958.2006.05182.x. PMID 16796673. S2CID 23738804.
- ^ Irving JA, Cabrita LD, Rossjohn J, Pike RN, Bottomley SP, Whisstock JC (April 2003). "The 1.5 A crystal structure of a prokaryote serpin: controlling conformational change in a heated environment". Structure. 11 (4): 387–397. doi:10.1016/S0969-2126(03)00057-1. PMID 12679017.
- ^ Fulton KF, Buckle AM, Cabrita LD, Irving JA, Butcher RE, Smith I, Reeve S, Lesk AM, Bottomley SP, Rossjohn J, Whisstock JC (March 2005). "The high resolution crystal structure of a native thermostable serpin reveals the complex mechanism underpinning the stressed to relaxed transition". The Journal of Biological Chemistry. 280 (9): 8435–8442. doi:10.1074/jbc.M410206200. PMID 15590653.
- ^ Turner PC, Moyer RW (September 2002). "Poxvirus immune modulators: functional insights from animal models". Virus Research. 88 (1–2): 35–53. doi:10.1016/S0168-1702(02)00119-3. PMID 12297326.
- ^ a b Richardson J, Viswanathan K, Lucas A (January 2006). "Serpins, the vasculature, and viral therapeutics". Frontiers in Bioscience. 11: 1042–1056. doi:10.2741/1862. PMID 16146796.
- ^ Jiang J, Arp J, Kubelik D, Zassoko R, Liu W, Wise Y, Macaulay C, Garcia B, McFadden G, Lucas AR, Wang H (November 2007). "Induction of indefinite cardiac allograft survival correlates with toll-like receptor 2 and 4 downregulation after serine protease inhibitor-1 (Serp-1) treatment". Transplantation. 84 (9): 1158–1167. doi:10.1097/01.tp.0000286099.50532.b0. PMID 17998872. S2CID 20168458.
- ^ Dai E, Guan H, Liu L, Little S, McFadden G, Vaziri S, Cao H, Ivanova IA, Bocksch L, Lucas A (May 2003). "Serp-1, a viral anti-inflammatory serpin, regulates cellular serine proteinase and serpin responses to vascular injury". The Journal of Biological Chemistry. 278 (20): 18563–18572. doi:10.1074/jbc.M209683200. PMID 12637546.
- ^ Turner PC, Sancho MC, Thoennes SR, Caputo A, Bleackley RC, Moyer RW (August 1999). "Myxoma virus Serp2 is a weak inhibitor of granzyme B and interleukin-1beta-converting enzyme in vitro and unlike CrmA cannot block apoptosis in cowpox virus-infected cells". Journal of Virology. 73 (8): 6394–6404. doi:10.1128/JVI.73.8.6394-6404.1999. PMC 112719. PMID 10400732.
- ^ Munuswamy-Ramanujam G, Khan KA, Lucas AR (December 2006). "Viral anti-inflammatory reagents: the potential for treatment of arthritic and vasculitic disorders". Endocrine, Metabolic & Immune Disorders Drug Targets. 6 (4): 331–343. doi:10.2174/187153006779025720. PMID 17214579.
- ^ Renatus M, Zhou Q, Stennicke HR, Snipas SJ, Turk D, Bankston LA, Liddington RC, Salvesen GS (July 2000). "Crystal structure of the apoptotic suppressor CrmA in its cleaved form". Structure. 8 (7): 789–797. doi:10.1016/S0969-2126(00)00165-9. PMID 10903953.
External links
[edit]- PDB Molecule of the Month Serpin
- Merops protease inhibitor claudication (Family I4) Archived 8 December 2016 at the Wayback Machine
- Serpins at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
- James Whisstock laboratory at Monash University
- Jim Huntington laboratory Archived 30 October 2016 at the Wayback Machine at University of Cambridge
- Frank Church laboratory at University of North Carolina at Chapel Hill
- Paul Declerck laboratory at Katholieke Universiteit Leuven
- Tom Roberts laboratory at University of Sydney
- Robert Fluhr laboratory at Weizmann Institute of Science
- Peter Gettins laboratory at University of Illinois at Chicago
- Overview of all the structural information available in the PDB for UniProt: P01009 (Human Alpha-1-antitrypsin) at the PDBe-KB.