List of taxa with candidatus status
This is a list of taxa with Candidatus status. For taxa not covered by this list, see also:
- the GenBank taxonomy for "effective" names as published;
- the Candidatus Lists and LPSN for latinate names, some sanitized to match the Code.[1]
Phyla[edit]
- "Ca. Absconditabacteria" (previously Candidate phylum SR1)[2][3][4][5]
- ABY1[6] aka OD1-ABY1, subgroup of OD1 ("Ca. Parcubacteria")[7]
- Candidate phylum AC1[4]
- "Ca. Acetothermia" (previously Candidate phylum OP1)[6][3][4][5]
- "Ca. Aerophobetes" (previously Candidate phylum CD12 or BHI80-139)[3][4]
- "Ca. Aminicenantes" (previously Candidate phylum OP8)[6][3][4][5]
- aquifer1[3]
- aquifer2[3]
- "Ca. Berkelbacteria" (previously Candidate phylum ACD58)[4][8]
- BRC1[6][4][5]
- CAB-I[4]
- "Ca. Calescamantes" (previously Candidate phylum EM19)[3][4][5]
- Candidate phylum CPR2[4]
- Candidate phylum CPR3[4]
- Candidate phylum NC10[6][4]
- Candidate phylum OP2[4]
- Candidate phylum RF3[4]
- Candidate phylum SAM[4]
- Candidate phylum SPAM[4]
- Candidate phylum TG2[4]
- Candidate phylum VC2[4]
- Candidate phylum WS1[4]
- Candidate phylum WS2[4][6]
- Candidate phylum WS4[4]
- Candidate phylum WYO[4]
- CKC4[3]
- "Ca. Cloacimonetes" (previously Candidate phylum WWE1)[3][5]
- CPR1[4]
- "Ca. Dependentiae" (previously Candidate phylum TM6)[3][4][6][9]
- EM 3[4]
- "Ca. Endomicrobia" Stingl et al. 2005[10]
- "Ca. Fermentibacteria" (Hyd24-12)[4][3][11]
- "Ca. Fervidibacteria" (previously Candidate phylum OctSpa1-106)[4]
- GAL08[3]
- GAL15[4]
- GN01[4]
- GN03[4]
- GN04[4]
- GN05[4]
- GN06[4]
- GN07[4]
- GN08[4]
- GN09[4]
- GN10[4]
- GN11[4]
- GN12[4]
- GN13[4]
- GN14[4]
- GN15[4]
- GOUTA4[3]
- "Ca. Gracilibacteria" (previously Candidate phylum GN02, BD1-5, or BD1-5 group)[6][3][4]
- Guaymas1[6]
- "Ca. Hydrogenedentes" (previously Candidate phylum NKB19)[6][3][4][5]
- JL-ETNP-Z39[3][4]
- "Ca. Katanobacteria" (previously Candidate phylum WWE3)[2][4]
- Kazan-3B-09[3]
- KD3-62[4]
- kpj58rc[4]
- KSA1[4]
- KSA2[4]
- KSB1[4]
- KSB2[4]
- KSB4[4]
- "Ca. Latescibacteria" (previously Candidate phylum WS3)[6][3][4][5]
- LCP-89[3]
- LD1-PA38[3]
- "Ca. Marinamargulisbacteria" (previously Candidate division ZB3)[12]
- "Ca. Marinimicrobia" (previously Marine Group A[6] or Candidate phylum SAR406)[3][4][5]
- "Ca. Melainabacteria"
- "Ca. Microgenomates" (previously Candidate phylum OP11)[6][3][4][5]
- "Ca. Modulibacteria" (previously Candidate phylum KSB3)[4][13]
- MSBL2[4]
- MSBL3[4]
- MSBL4[4]
- MSBL5[4]
- MSBL6[4]
- NPL-UPA2[4]
- NT-B4[4]
- OC31[3]
- "Ca. Omnitrophica" (previously Candidate phylum OP3)[6][3][4][5]
- OP4[4]
- OP6[4]
- OP7[4]
- OS-K/OS-K group[6][4]
- "Ca. Parcubacteria" (previously Candidate phylum OD1)[3][4][5]
- PAUC34f[3]
- "Ca. Peregrinibacteria" (previously Candidate phylum PER)[4][14][15][16][17]
- "Ca. Poribacteria"[4][5][18]
- RsaHF231[3]
- S2R-29[3]
- "Ca. Saccharibacteria" (previously Candidate phylum TM7)[3][4][5][19][6][20][21]
- SBR1093[6][4]
- SBYG-2791[3]
- Candidate phylum SC3[6]
- Candidate phylum SC4[6]
- Sediment-1[4]
- Sediment-2[4]
- Sediment-3[4]
- Sediment-4[4]
- SHA-109[3]
- SM2F11[3]
- TA06[3][4]
- "Ca. Tectomicrobia"[22]
- TG3 ("Ca. Chitinivibrionia")
- WCHB1-60[3]
- WD272[3]
- WOR-1[4]
- WOR-3[4]
- WPS-1[4][5]
- WPS-2[4][5]
- WS5[6][4]
- WS6[6][3][4][23]
- ZB2 aka OD1-ZB2 subgroup of OD1 ("Ca. Parcubacteria")[7]
- "Ca. Zixibacteria"[4]
Classes[edit]

- "Ca. Lambdaproteobacteria" Anantharaman et al. 2016
- "Ca. Muproteobacteria" Anantharaman et al. 2016
- "Ca. Zetaproteobacteria" Emerson et al. 2007[10]
Orders[edit]
- "Ca. Nitrosopumilales" Könneke et al. 2005[10]
Families[edit]
- "Ca. Midichloriaceae" Montagna et al. 2013[10]
- "Ca. Nitrosopumilaceae" Könneke et al. 2005[10]
- "Ca. Procabacteriaceae"
- "Ca. Rhabdochlamydiaceae"
Genera[edit]
- "Ca. Accumulibacter" Hesselmann et al. 1999[10]
- "Ca. Allobeggiatoa" Hinck et al. 2011[10]
- "Ca. Arthromitus" Snel et al. 1995[10]
- "Ca. Bacilloplasma" Kostanjšek et al. 2007[10]
- "Ca. Baumannia" Moran et al. 2003[10]
- "Ca. Blochmannia" Sauer et al. 2000[10]
- "Ca. Carsonella" Thao et al. 2000[10]
- "Ca. Chlorothrix" Klappenbach and Pierson 2004[10]
- "Ca. Competibacter" Crocetti et al. 2002[10]
- "Ca. Contubernalis" Zhilina et al. 2005[10]
- "Ca. Desulforudis"
- "Ca. Endomicrobium" Stingl et al. 2005[10]
- "Ca. Epixenosoma"
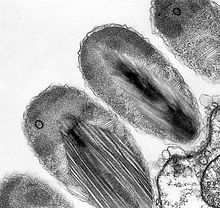
Transmission electron micrograph of stage II epixenosomes. - "Ca. Giganthauma"
- "Ca. Glomeribacter" Bianciotto et al. 2003[10]
- "Ca. Kleidoceria" Kuechler et al. 2010[10]
- "Ca. Kuenenia" Schmid et al. 2000[10]
- "Ca. Liberibacter"
- "Ca. Lokiarchaeum"
- "Ca. Lumbricincola" Nechitaylo et al. 2009[10]
- "Ca. Magnetobacterium" Murray and Stackebrandt 1995[10]
- "Ca. Methanoregula"
- "Ca. Microthrix"
- "Ca. Nardonella" Lefèvre et al. 2004[10]
- "Ca. Nitrosoarchaeum" Blainey et al. 2011[10]
- "Ca. Nitrosopumilus" Könneke et al. 2005[10]
- "Ca. Pelagibacter" Rappe et al. 2002[10]
- "Ca. Photodesmus" Hendry and Dunlap 2011[10]
- "Ca. Phytoplasma" Firrao et al. 2004[10]
- "Ca. Portiera" Thao and Baumann 2004[10]
- "Ca. Procabacter" Horn et al. 2002[10]
- "Ca. Rhabdochlamydia"
- "Ca. Riegeria" Gruber-Vodicka et al. 2011[10]
- "Ca. Rohrkolberia" Kuechler et al. 2011[10]
- "Ca. Salinibacter" Antón et al. 2000[10]
- "Ca. Savagella" Thompson et al. 2012[10]
- "Ca. Scalindua"
- "Ca. Similichlamydia" Stride et al. 2013[10]
- "Ca. Symbiothrix" Hongoh et al. 2007[10]
- "Ca. Tremblaya" Thao et al. 2002[10]
- "Ca. Xiphinematobacter" Vandekerckhove et al. 2000[10]
Species and subspecies[edit]
Unless otherwise noted (♦), these entries are from LPSN.[10]
- "Ca. Acaryochloris bahamiensis" López-Legentil et al. 2011
- "Ca. Acidianus copahuensis" Giaveno et al. 2013
- "Ca. Aciduliprofundum boonei" Schouten et al. 2008

- "Ca. Accumulibacter phosphatis" Hesselmann et al. 1999
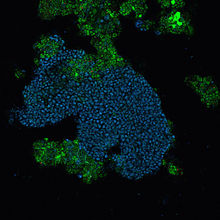
A FISH image of a floc from a lab-scale EBPR reactor. The PAOmix probe (Cy5) was used to stain "Ca. Accumulibacter phosphatis", which appear in blue - "Ca. Actinobaculum timonae" Drancourt et al. 2004
- "Ca. Adiaceo aphidicola" Darby et al. 2005
- "Ca. Allobeggiatoa salina" Hinck et al. 2011
- "Ca. Alysiomicrobium bavaricum" Kragelund et al. 2006
- "Ca. Alysiosphaera europaea" Kragelund et al. 2006
- "Ca. Amoebinatus massiliae" Greub et al. 2004
- "Ca. Amoebophilus asiaticus" Horn et al. 2001
- "Ca. Amphibiichlamydia ranarum" Martel et al. 2013
- "Ca. Anadelfobacter veles" Vannini et al. 2010
- "Ca. Ancillula trichonymphae" Strassert et al. 2012
- "Ca. Anammoxoglobus propionicus" Kartal et al. 2007
- "Ca. Aquiluna rubra" Hahn 2009
- "Ca. Aquirestis calciphila" Hahn and Schauer 2007
- "Ca. Arcobacter sulfidicus" Wirsen et al. 2002
- "Ca. Arsenophonus arthropodicus" Dale et al. 2002
- "Ca. Arsenophonus phytopathogenicus" Bressan et al. 2012
- "Ca. Arsenophonus triatominarum" Hypsa and Dale 1997
- "Ca. Aschnera chinzeii" Hosokawa et al. 2012
- "Ca. Atelocyanobacterium thalassa(♦)
- "Ca. Azoamicus ciliaticola" Graf et al. 2021[24]
- "Ca. Bacteroides massiliae" Drancourt et al. 2004
- "Ca. Bartonella ancashi" Blazes et al. 2013
- "Ca. Bartonella antechini" Kaewmongkol et al. 2011
- "Ca. Bartonella breitschwerdtii(♦)
- "Ca. Bartonella bandicootii" Kaewmongkol et al. 2011
- "Ca. Bartonella durdenii(♦)
- "Ca. Bartonella eldjazairii(♦)
- "Ca. Bartonella mayotimonensis" Lin et al. 2010
- "Ca. Bartonella melophagi(♦)
- "Ca. Bartonella merieuxii" Chomel et al. 2012
- "Ca. Bartonella monaxi(♦)
- "Ca. Bartonella rudakovii(♦)
- "Ca. Bartonella thailandensis" Saisongkorh et al. 2009
- "Ca. Bartonella volans(♦)
- "Ca. Bartonella woyliei" Kaewmongkol et al. 2011
- "Ca. Baumannia cicadellinicola" Moran et al. 2003
- "Ca. Benitsuchiphilus tojoi" Hosokawa et al. 2010
- "Ca. Berkiella aquae" Mehari et al. 2016
- "Ca. Berkiella cookevillensis" Mehari et al. 2016
- "Ca. Blochmannia floridanus" Sauer et al. 2000
- "Ca. Blochmannia herculeanus" Sauer et al. 2000
- "Ca. Blochmannia rufipes" Sauer et al. 2000
- "Ca. Borrelia tachyglossi" Loh et al. 2017
- "Ca. Borrelia texasensis" Lin et al. 2005
- "Ca. Branchiomonas cysticola" Toenshoff et al. 2012
- "Ca. Brocadia anammoxidans" Jetten et al. 2001
- "Ca. Brocadia caroliniensis" Magrí et al. 2012
- "Ca. Brocadia fulgida" Kartal et al. 2004
- "Ca. Brocadia sinica" Hu et al. 2010
- "Ca. Brownia rhizoecola" Gruwell et al. 2010
- "Ca. Burkholderia andongensis" Lemaire et al. 2011
- "Ca. Burkholderia calva" Van Oevelen et al. 2004
- "Ca. Burkholderia crenata(♦)
- "Ca. Burkholderia hispidae" Lemaire et al. 2012
- "Ca. Burkholderia kirkii" Van Oevelen et al. 2002
- "Ca. Burkholderia mamillata(♦)
- "Ca. Burkholderia nigropunctata" Van Oevelen et al. 2004
- "Ca. Burkholderia petitii" Lemaire et al. 2011
- "Ca. Burkholderia rigidae" Lemaire et al. 2012
- "Ca. Burkholderia schumannianae" Lemaire et al. 2012
- "Ca. Burkholderia verschuerenii(♦)
- "Ca. Burkholderia virens(♦)
- "Ca. Caedibacter acanthamoebae" Horn et al. 1999
- "Ca. Caldiarchaeum subterraneum" Nunoura et al. 2011
- "Ca. Campylobacter hominis" Lawson et al. 1998
- "Ca. Captivus acidiprotistae" Baker et al. 2003
- "Ca. Cardinium hertigii" Zchori-Fein et al. 2004
- "Ca. Carsonella ruddii" Thao et al. 2000
- "Ca. Chloracidobacterium thermophilum" Bryant et al. 2007
- "Ca. Chlorothrix halophila" Klappenbach and Pierson 2004
- "Ca. Chryseobacterium massiliae" Greub et al. 2004
- "Ca. Chryseobacterium timonae" Drancourt et al. 2004
- "Ca. Clavochlamydia salmonicola" Karlsen et al. 2008
- "Ca. Cloacamonas acidaminovorans" Pelletier et al. 2008
- "Ca. comitans" Jacobi et al. 1996
- "Ca. Competibacter phosphatis" Crocetti et al. 2002
- "Ca. Consessoris aphidicola" Darby et al. 2005
- "Ca. Contubernalis alkalaceticum" Zhilina et al. 2005
- "Ca. Cryptoprodotis polytropus" Ferrantini et al. 2009
- "Ca. Curculioniphilus buchneri" Toju et al. 2009
- "Ca. Cyrtobacter comes" Vannini et al. 2010
- "Ca. Cyrtobacter zanobii" Boscaro et al. 2013
- "Ca. Defluviella procrastinata" Boscaro et al. 2013
- "Ca. Desulforudis audaxviator" Chivian et al. 2008

a colony of "Ca. Desulforudis audaxviator" discovered in a gold mine near Johannesburg, South Africa. - "Ca. Desulfovibrio trichonymphae" Sato et al. 2009
- "Ca. Devosia euplotis" Vannini et al. 2004
- "Ca. Ecksteinia adelgidicola" Toenshoff et al. 2012
- "Ca. Ehrlichia walkerii" Brouqui et al. 2003
- "Ca. Endobugula glebosa" Lim and Haygood 2004
- "Ca. Endobugula sertula" Haygood and Davidson 1997
- "Ca. Endolissoclinum faulkneri" Kwan et al. 2012
- "Ca. Endoecteinascidia frumentensis" Pérez-Matos et al. 2007
- "Ca. Endomicrobium pyrsonymphae" Stingl et al. 2005
- "Ca. Endomicrobium trichonymphae" Stingl et al. 2005
- "Ca. Endonucleobacter bathymodioli" Zielinski et al. 2009
- "Ca. Endoriftia persephone" Robidart et al. 2008
- "Ca. Endowatersipora palomitas" Anderson and Haygood 2007
- "Ca. Endowatersipora rubus" Anderson and Haygood 2007
- "Ca. Entotheonella palauensis" Schmidt et al. 2000
- "Ca. Epiflobacter spp." Xia et al. 2008
- "Ca. Erwinia dacicola" Capuzzo et al. 2005
- "Ca. Evansia muelleri" Kuechler et al. 2013
- "Ca. Flaviluna lacus" Hahn 2009
- "Ca. Fodinabacter communificans" Bertin et al. 2011
- "Ca. Francisella noatunensis subsp. endociliophora" Schrallhammer et al. 2011
- "Ca. Frankia datiscae" Persson et al. 2011
- "Ca. Fritschea bemisiae" Everett et al. 2005
- "Ca. Fritschea eriococci" Everett et al. 2005
- "Ca. Gillettellia cooleyia" Toenshoff et al. 2012
- "Ca. Gilliamella apicola" Martinson et al. 2012
- "Ca. Glomeribacter gigasporarum" Bianciotto et al. 2003
- "Ca. Gortzia infectiva" Boscaro et al. 2012
- "Ca. Haliscomenobacter calcifugiens" Hahn and Schauer 2007
- "Ca. Halomonas phosphatis" Nguyen et al. 2012
- "Ca. Hamiltonella defensa" Moran et al. 2005
- "Ca. Helicobacter bovis" De Groote et al. 1999
- "Ca. Helicobacter heilmannii" O'Rourke et al. 2004
- "Ca. Helicobacter suis" De Groote et al. (includes strains previously known as Helicobacter heilmannii type one)
- "Ca. Heliomonas lunata" Asao et al. 2012
- "Ca. Hemipteriphilus asiaticus" Bing et al. 2013
- "Ca. Hemobacterium ranarum" Zhang and Rikihisa 2004 (previously known as Aegyptianella ranarum)
- "Ca. Hepatincola porcellionum" Wang et al. 2004
- "Ca. Hepatobacter penaei" Numan et al. 2013
- "Ca. Hepatoplasma crinochetorum" Wang et al. 2004
- "Ca. Hodgkinia cicadicola" McCutcheon et al. 2009
- "Ca. Holdemania massiliensis(♦)
- "Ca. Ishikawaella capsulata" Hosokawa et al. 2006
- "Ca. Isobeggiatoa divolgata" Salman et al. 2011
- "Ca. Jettenia asiatica" Quan et al. 2008
- "Ca. Kinetoplastibacterium blastocrithidii" Du et al. 1994
- "Ca. Kinetoplastibacterium crithidii" Du et al. 1994
- "Ca. Kleidoceria schneideri" Kuechler et al. 2010
- "Ca. Kopriimonas aquarianus" Quinn et al. 2012
- "Ca. Korarchaeum cryptofilum" Elkins et al. 2008
- "Ca. Koribacter versatilis" Ward et al. 2009
- "Ca. Kuenenia stuttgartiensis" Schmid et al. 2000
- "Ca. Lariskella arthropodarum" Matsuura et al. 2012
- "Ca. Legionella jeonii" Park et al. 2004
- "Ca. Liberibacter americanus" Teixeira et al. 2005 - one cause of Citrus greening disease

Citrus greening disease on mandarin oranges - "Ca. Liberibacter africanus" Roberts et al. 2015 - one cause of Citrus greening disease
- "Ca. Liberibacter africanus" corrig. Jagoueix et al. 1994 (previously "Ca. Liberobacter africanum)
- "Ca. Liberibacter africanus subsp. capensis" Garnier et al. 2000
- "Ca. Liberibacter asiaticus" corrig. Jagoueix et al. 1994 (previously "Ca. Liberobacter asiaticum) - one cause of Citrus greening disease
- "Ca. Liberibacter europaeus" Raddadi et al. 2011
- "Ca. Liberibacter psyllaurous" Hansen et al. 2008
- "Ca. Liberibacter solanacearum" Liefting et al. 2009 - cause of Psyllid yellows and Zebra chip

Potato chips exhibiting Zebra chip - "Ca. Limnoluna rubra" Hahn 2009
- "Ca. Macropleicola appendiculatae" Kölsch et al. 2009
- "Ca. Macropleicola muticae" Kölsch et al. 2009
- "Ca. Magnetobacterium bavaricum" Spring et al. 1993
- "Ca. Magnetoglobus multicellularis" Abreu et al. 2007
- "Ca. Magnospira bakii" Snaidr et al. 1999
- "Ca. Maribeggiatoa vulgaris" Salman et al. 2011
- "Ca. Marispirochaeta associata" Shivani et al. 2016
- "Ca. Marithioploca araucae" Salman et al
- "Ca. Marithrix sessilis" Salman et al. 2011
- "Ca. Mesochlamydia elodeae" Corsaro et al. 2013
- "Ca. Metachlamydia lacustris" Corsaro et al. 2010
- "Ca. Methanogranum caenicola" Iino et al. 2013
- "Ca. Methanomethylophilus alvus" Borrel et al. 2012
- "Ca. Methanoregula boonei" Bräuer et al. 2006
- "Ca. Methylacidiphilum infernorum" Hou et al. 2008
- "Ca. Methylomirabilis oxyfera" Ettwig et al. 2010
- "Ca. Micrarchaeum acidiphilum(♦)
- "Ca. Microthrix calida" Levantesi et al. 2006
- "Ca. Microthrix parvicella" Blackall et al. 1996
- "Ca. Midichloria mitochondrii" Sassera et al. 2006
- "Ca. Monilibacter batavus" Kragelund et al. 2006
- "Ca. Moranella endobia" McCutcheon and von Dohlen 2011
- "Ca. Mycoplasma aoti" Barker et al. 2011
- "Ca. Mycoplasma corallicola" Neulinger et al. 2009
- "Ca. Mycoplasma haematoparvum" Sykes et al. 2005
- "Ca. Mycoplasma haemobos" Tagawa et al. 2008
- "Ca. Mycoplasma haemodidelphidis" Messick et al. 2002
- "Ca. Mycoplasma haemofelis" Neimark et al. 2001 (synonymous with Haemobartonella felis)

"Ca. Mycoplasma haemofelis" with Wright-Giemsa stain - "Ca. Mycoplasma haemolamae" Messick et al. 2002
- "Ca. Mycoplasma haemomacaque" Maggi et al. 2013
- "Ca. Mycoplasma haemominutum" Foley and Pedersen 2001 (previously "Haemobartonella felis small form (Hfsm)")
- "Ca. Mycoplasma haemomuris" Neimark et al. 2001 (synonymous with Haemobartonella muris)
- "Ca. Mycoplasma haemomuris subsp. musculi" Harasawa et al. 2015
- "Ca. Mycoplasma haemomuris subsp. ratti" Harasawa et al. 2015
- "Ca. Mycoplasma haemosuis" Neimark et al. 2001 (synonymous with Eperythrozoon suis and Mycoplasma suis)
- "Ca. Mycoplasma haemovis" Hornok et al. 2009
- "Ca. Mycoplasma haemozalophi" Volokhov et al. 2011
- "Ca. Mycoplasma kahanei" Neimark et al. 2002
- "Ca. Mycoplasma ravipulmonis" Neimark et al. 1998
- "Ca. Mycoplasma turicensis" Willi et al. 2006
- "Ca. Mycoplasma wenyonii" Neimark et al. 2001 (synonymous with Eperythrozoon wenyonii)
- "Ca. Nebulobacter yamunensis" Boscaro et al. 2012
- "Ca. Neoehrlichia arcana" Gofton et al. 2016
- "Ca. Neoehrlichia australis" Gofton et al. 2016
- "Ca. Neoehrlichia lotoris" Yabsley et al. 2008
- "Ca. Neoehrlichia mikurensis" Kawahara et al. 2004
- "Ca. Nitrosoarchaeum koreensis" Kim et al. 2011
- "Ca. Nitrosoarchaeum limnia" Blainey et al. 2011
- "Ca. Nitrosocaldus yellowstonii" de la Torre et al. 2008
- "Ca. Nitrosopelagicus brevis(♦)
- "Ca. Nitrosopumilus koreensis" Park et al. 2012
- "Ca. Nitrosopumilus maritimus" Könneke et al. 2005
- "Ca. Nitrosopumilus salaria" Mosier et al. 2012
- "Ca. Nitrosopumilus sediminis" Park et al. 2012
- "Ca. Nitrososphaera gargensis" Hatzenpichler et al. 2008
- "Ca. Nitrososphaera viennensis" Tourna et al. 2011
- "Ca. Nitrospira bockiana" Lebedeva et al. 2008
- "Ca. Nitrospira defluvii" Spieck et al. 2006
- "Ca. Nitrosotalea devanaterra" Lehtovirta-morley et al. 2011
- "Ca. Nitrotoga arctica" Alawi et al. 2007
- "Ca. Nostocoida limicola" Blackall et al. 2000 (strains Ben 17, Ben 18, Ben 67, Ben 68 and Ben 74 are now Tetrasphaera jenkinsii, strain Ben 70 is Tetrasphaera vanveenii, and strains Ver 1 and Ver 2 are Tetrasphaera veronensis)
- "Ca. Odyssella thessalonicensis" Birtles et al. 2000
- "Ca. Ovobacter propellens" Fenchel and Thar 2004
- "Ca. Paenicardinium endonii" Noel and Atibalentja 2006
- "Ca. Parabeggiatoa communis" Salman et al. 2011
- "Ca. Paracaedibacter acanthamoebae" Horn et al. 1999
- "Ca. Paracaedibacter symbiosus" Horn et al. 1999
- "Ca. Paraholospora nucleivisitans" Eschbach et al. 2009
- "Ca. Parilichlamydia carangidicola" Stride et al. 2013
- "Ca. Parvarchaeum acidophilus(♦)
- "Ca. Pasteuria aldrichii" Giblin-Davis et al. 2011
- "Ca. Pasteuria usgae" Giblin-Davis et al. 2003
- "Ca. Pelagibacter ubique" Rappe et al. 2002
- "Ca. Peptostreptococcus massiliae" Drancourt et al. 2004
- "Ca. Phlomobacter fragariae" Zreik et al. 1998
- "Ca. Photodesmus katoptron" Hendry and Dunlap 2011
- "Ca. Phytoplasma 16SrII-U" Yang et al. 2016
- "Ca. Phytoplasma allocasuarinae" Marcone et al. 2004
- "Ca. Phytoplasma americanum" Lee et al. 2006
- "Ca. Phytoplasma asteris" Lee et al. 2004
- "Ca. Phytoplasma aurantifolia" Zreik et al. 1995
- "Ca. Phytoplasma australiense" Davis et al. 1997
- "Ca. Phytoplasma balanitae" Win et al. 2013
- "Ca. Phytoplasma brasiliense" Montano et al. 2001
- "Ca. Phytoplasma caricae" Arocha et al. 2005
- "Ca. Phytoplasma castaneae" Jung et al. 2002
- "Ca. Phytoplasma cirsii" Safarova et al. 2016
- "Ca. Phytoplasma convolvuli" Martini et al. 2012
- "Ca. Phytoplasma costaricanum" Lee et al. 2011
- "Ca. Phytoplasma cynodontis" Marcone et al. 2004
- "Ca. Phytoplasma fragariae" Valiunas et al. 2006
- "Ca. Phytoplasma fraxini" Griffiths et al. 1999 - cause of Lilac witches'-broom

Symptoms of ash yellows disease caused by "Ca. Phytoplasma fraxini" on green ash - "Ca. Phytoplasma graminis" Arocha et al. 2005
- "Ca. Phytoplasma hispanicum" Davis et al. 2016
- "Ca. Phytoplasma japonicum" Sawayanagi et al. 1999
- "Ca. Phytoplasma lycopersici" Arocha et al. 2007
- "Ca. Phytoplasma malaysianum" Nejat et al. 2013
- "Ca. Phytoplasma mali" Seemüller and Schneider 2004
- "Ca. Phytoplasma meliae" Fernandez et al. 2016
- "Ca. Phytoplasma omanense" Al-Saady et al. 2008
- "Ca. Phytoplasma oryzae" Jung et al. 2003
- "Ca. Phytoplasma palmicola" Harrison et al. 2014
- "Ca. Phytoplasma phoenicium" Verdin et al. 2003
- "Ca. Phytoplasma pini" Schneider et al. 2005
- "Ca. Phytoplasma pruni" Davis et al. 2013
- "Ca. Phytoplasma prunorum" Seemüller and Schneider 2004
- "Ca. Phytoplasma pyri" Seemüller and Schneider 2004
- "Ca. Phytoplasma rhamni" Marcone et al. 2004
- "Ca. Phytoplasma rubi" Malembic-Maher et al. 2011
- "Ca. Phytoplasma rubi" Franova et al. 2016
- "Ca. Phytoplasma solani" Quaglino et al. 2013
- "Ca. Phytoplasma spartii" Marcone et al. 2004
- "Ca. Phytoplasma sudamericanum" Davis et al. 2012
- "Ca. Phytoplasma tamaricis" Zhao et al
- "Ca. Phytoplasma trifolii" Hiruki and Wang 2004
- "Ca. Phytoplasma ulmi" Lee et al. 2004
- "Ca. Phytoplasma vitis" Marzorati et al. 2006 - cause of Flavescence dorée

Flavescence dorée on grapevine - "Ca. Phytoplasma ziziphi" Jung et al. 2003
- "Ca. Piscichlamydia salmonis" Draghi et al. 2004
- "Ca. Planktoluna difficilis" Hahn 2009
- "Ca. Planktomarina temperata" Giebel et al. 2011
- "Ca. Planktophila limnetica" Jezbera et al. 2009
- "Ca. Portiera aleyrodidarum" Thao and Baumann 2004
- "Ca. Prevotella massiliensis" Drancourt et al. 2004
- "Ca. Procabacter acanthamoebae" Horn et al. 2002
- "Ca. Profftia tarda" Toenshoff et al. 2012
- "Ca. Profftia virida" Toenshoff et al. 2012
- "Ca. Protochlamydia amoebophila" Collingro et al. 2005
- "Ca. Puchtella pedicinophila" Fukatsu et al. 2009
- "Ca. Puniceispirillum marinum" Oh et al. 2010
- "Ca. Purcelliella pentastirinorum" Bressan et al. 2009
- "Ca. Regiella insecticola" Moran et al. 2005
- "Ca. Renichlamydia lutjani" Corsaro and Work 2012
- "Ca. Rhabdochlamydia crassificans" Corsaro et al. 2006
- "Ca. Rhabdochlamydia porcellionis" Kostanjšek (morphology matches the description of Chlamydia isopodii)
- "Ca. Rhizobium massiliae" Greub et al. 2004
- "Ca. Rhodoluna limnophila" Hahn 2009
- "Ca. Rhodoluna planktonica" Hahn 2009
- "Ca. Rickettsia amblyommii" Labruna et al. 2004
- "Ca. Rickettsia andeanae" Jiang et al. 2005
- "Ca. Rickettsia asemboensis" Jiang et al. 2013
- "Ca. Rickettsia barbariae" Mura et al. 2008
- "Ca. Rickettsia hebeiii" Yaxue et al. 2011
- "Ca. Rickettsia hungarica" Hornok et al. 2010
- "Ca. Rickettsia hoogstraalii" Mattila et al. 2007[25]
- "Ca. Rickettsia kellyi" Rolain et al. 2006
- "Ca. Rickettsia kingi" Anstead and Chilton 2013
- "Ca. Rickettsia kotlanii" Sréter-Lancz et al. 2006
- "Ca. Rickettsia liberiensis" Mediannikov et al. 2012
- "Ca. Rickettsia principis" Mediannikov et al. 2006
- "Ca. Rickettsia senegalensis" Mediannikov, Aubadie-Ladrix, and Raoult 2015[26]
- "Ca. Rickettsia tarasevichiae" Shpynov et al. 2003
- "Ca. Rickettsia tasmanensis" Izzard et al. 2009
- "Ca. Rickettsia vini" Palomar et al. 2012
- "Ca. Riegeria galateiae" Gruber-Vodicka et al. 2011
- "Ca. Riesia pediculicola" Sasaki-Fukatsu et al. 2006
- "Ca. Rohrkolberia cinguli" Kuechler et al. 2011
- "Ca. Roseomonas massiliae" Greub et al. 2004
- "Ca. Ruthia magnifica" Newton et al. 2007
- "Ca. Scalindua arabica" Woebken et al. 2008
- "Ca. Scalindua brodae" Schmid et al. 2003
- "Ca. Scalindua pacifica" Dang et al. 2003
- "Ca. Scalindua profunda" Van De Vossenberg et al. 2008
- "Ca. Scalindua richardsii" Fuchsman et al. 2012
- "Ca. Scalindua sorokinii" Kuypers et al. 2003
- "Ca. Scalindua wagneri" Schmid et al. 2003
- "Ca. Serratia symbiotica" Moran et al. 2005
- "Ca. Similichlamydia latridicola" Stride et al. 2013
- "Ca. Snodgrassella alvi" Martinson et al. 2012
- "Ca. Snodgrassella alvi" Martinson et al. 2012
- "Ca. Sodalis melophagi" Chrudimský et al. 2012
- "Ca. Sodalis pierantonius" Oakeson et al. 2014
- "Ca. Sphaeronema italicum" Kragelund et al. 2006
- "Ca. Stammerula tephritidis" Mazzon et al. 2008
- "Ca. Steffania adelgidicola" Toenshoff et al. 2012
- "Ca. Streptomyces philanthi" Kaltenpoth et al. 2006
- "Ca. Sulcia muelleri" Moran et al. 2005
- "Ca. Sulfurovum sediminum" Park et al. 2012
- "Ca. Symbiothrix dinenymphae" Hongoh et al. 2007
- "Ca. Synechococcus spongiarum" Usher et al. 2004
- "Ca. Tammella caduceiae" Hongoh et al. 2007
- "Ca. Tenderia electrophaga" Eddie et al. 2016
- "Ca. Thermochlorobacter aerophilum" Liu et al. 2012
- "Ca. Thiobios zoothamnicoli" Rinke et al. 2006
- "Ca. Thiodictyon syntrophicum" Peduzzi et al. 2012
- "Ca. Thioglobus singularis" Marshall and Morris 2013
- "Ca. Thiomargarita joergensenii" Salman et al. 2011
- "Ca. Thiomargarita nelsonii" Salman et al. 2011
- "Ca. Thiophysa hinzei" Salman et al. 2011
- "Ca. Thiopilula aggregata" Salman et al. 2011
- "Ca. Thioturbo danicus" Muyzer et al. 2005
- "Ca. Tremblaya phenacola" Gruwell et al. 2010
- "Ca. Tremblaya princeps" Thao et al. 2002
- "Ca. Troglogloea absoloni" Kostanjšek et al. 2013
- "Ca. Uzinura diaspidicola" Gruwell et al. 2007
- "Ca. Vallotia cooleyia" Toenshoff et al. 2012
- "Ca. Vallotia tarda" Toenshoff et al. 2012
- "Ca. Vallotia virida" Toenshoff et al. 2012
- "Ca. Veillonella atypica" Drancourt et al. 2004
- "Ca. Vesicomyosocius okutanii" Kuwahara et al. 2007
- "Ca. Vestibaculum illigatum" Stingl et al. 2004
- "Ca. Vidania fulgoroideae" Gonella et al. 2011
- "Ca. Wolinella africanus" Bohr et al. 2003
- "Ca. Xenohaliotis californiensis" Friedman et al. 2000
- "Ca. Xiphinematobacter americani" Vandekerckhove et al. 2000
- "Ca. Xiphinematobacter brevicolli" Vandekerckhove et al. 2000
- "Ca. Xiphinematobacter rivesi" Vandekerckhove et al. 2000
- "Ca. Zinderia insecticola" McCutcheon and Moran 2010
Former candidatus taxa[edit]
- Armatimonadota, formerly Candidate phylum OP10[6][27][28]
- Atribacterota (previously Candidate phylum OP9 or JS1)[3][4][5][2][6]
- Caldisericota, formerly Candidate phylum OP5[6][29]
- Elusimicrobiota, formerly Termite Group 1[30]
- Ignavibacteriota (previously Candidate phylum ZB1)[5][31]
- Lentisphaerota, formerly vadinBE97[6][32]
- Nitrospinota[4][5][33]
See also[edit]
- Uncultivated bacterial phyla and metagenomics
- Open nomenclature, a system of notations used in taxonomy to indicate a taxonomist's judgement about taxon affinities
- Glossary of scientific naming
- incertae sedis, a taxon of uncertain position in a classification
- nomen dubium, a name of unknown or doubtful application
References[edit]
- ^ Oren, A (November 2021). "Nomenclature of prokaryotic 'Candidatus' taxa: establishing order in the current chaos". New Microbes and New Infections. 44: 100932. doi:10.1016/j.nmni.2021.100932. PMC 8487987. PMID 34631108.
- ^ a b c Hug LA; et al. (2016). "A new view of the tree of life". Nature Microbiology. Article 16048 (5): 16048. doi:10.1038/nmicrobiol.2016.48. PMID 27572647.
- ^ a b c d e f g h i j k l m n o p q r s t u v w x y z aa ab ac ad ae af ag ah ai aj ak "ARB-Silva: comprehensive ribosomal RNA database". The ARB development Team. Retrieved January 2, 2016.
- ^ a b c d e f g h i j k l m n o p q r s t u v w x y z aa ab ac ad ae af ag ah ai aj ak al am an ao ap aq ar as at au av aw ax ay az ba bb bc bd be bf bg bh bi bj bk bl bm bn bo bp bq br bs bt bu bv bw bx by bz ca cb cc cd ce cf cg ch ci cj "Taxonomy Browser". NCBI. Retrieved 2 January 2016.
- ^ a b c d e f g h i j k l m n o p q r s "Hierarchy Browser". Ribosomal database project. Archived from the original on November 21, 2008. Retrieved January 2, 2016.
- ^ a b c d e f g h i j k l m n o p q r s t u v w x y Rappé, Michael S.; Giovannoni, Stephen J. (2003). "The Uncultured Microbial Majority". Annual Review of Microbiology. 57: 369–94. doi:10.1146/annurev.micro.57.030502.090759. PMID 14527284.
- ^ a b Kenly A. Hiller, Kenneth H. Foreman, David Weisman, Jennifer L. Bowen: Permeable Reactive Barriers Designed To Mitigate Eutrophication Alter Bacterial Community Composition and Aquifer Redox Conditions. In: Appl Environ Microbiol v.81(20); 2015 Oct; pp.7114–7124. doi:10.1128/AEM.01986-15. PMC 4579450. PMID 26231655.
- ^ Wrighton KC; et al. (2014). "Metabolic interdependencies between phylogenetically novel fermenters and respiratory organisms in an unconfined aquifer". The ISME Journal. 8 (7): 1452–1463. doi:10.1038/ismej.2013.249. PMC 4069391. PMID 24621521.
- ^ Yeoh YK; et al. (2015). "Comparative Genomics of Candidate Phylum TM6 Suggests That Parasitism Is Widespread and Ancestral in This Lineage". Mol Biol Evol. 33 (4): 915–927. doi:10.1093/molbev/msv281. PMC 4776705. PMID 26615204.
- ^ a b c d e f g h i j k l m n o p q r s t u v w x y z aa ab ac ad ae af ag ah ai aj ak al Parte, A.C; Euzéby, J.P. (2017). "Names included in the category Candidatus". List of Prokaryotic names with Standing in Nomenclature. Retrieved 4 January 2018.
- ^ Kirkegaard, Rasmus Hansen; Dueholm, Morten Simonsen; McIlroy, Simon Jon; Nierychlo, Marta; Karst, Søren Michael; Albertsen, Mads; Nielsen, Per Halkjær (2016-10-01). "Genomic insights into members of the candidate phylum Hyd24-12 common in mesophilic anaerobic digesters". The ISME Journal. 10 (10): 2352–2364. doi:10.1038/ismej.2016.43. ISSN 1751-7370. PMC 5030696. PMID 27058503.
- ^ NCBI: Candidate division ZB3 (class), "Cyanobacteria"/"Melainabacteria" group
- ^ Sekiguchi Y; et al. (2015). "First genomic insights into members of a candidate bacterial phylum responsible for wastewater bulking". PeerJ. 3: e740. doi:10.7717/peerj.740. PMC 4312070. PMID 25650158.
- ^ Christopher T. Brown, Laura A. Hug, Brian C. Thomas et al.; et al. (2015). "Unusual biology across a group comprising more than 15% of domain Bacteria" (PDF). Nature. 523 (7559): 208–11. Bibcode:2015Natur.523..208B. doi:10.1038/nature14486. OSTI 1512215. PMID 26083755. S2CID 4397558.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ NCBI: Candidatus Peregrinibacteria (phylum)
- ^ UniProt: Taxonomy - Candidatus Peregrinibacteria (PHYLUM)
- ^ Karthik Anantharaman, Christopher T. Brown, David Burstein, Cindy Castelle: Analysis of five complete genome sequences for members of the class Peribacteria in the recently recognized Peregrinibacteria bacterial phylum. In: PeerJ 4(8):e1607; Jan 2016; doi:10.7717/peerj.1607
- ^ Fieseler L; et al. (2004). "Discovery of the Novel Candidate Phylum "Poribacteria" in Marine Sponges". Applied and Environmental Microbiology. 70 (6): 3724–32. Bibcode:2004ApEnM..70.3724F. doi:10.1128/AEM.70.6.3724-3732.2004. PMC 427773. PMID 15184179.
- ^ Albertsen M; et al. (2013). "Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes". Nat. Biotechnol. 31 (6): 533–8. doi:10.1038/nbt.2579. PMID 23707974. S2CID 11038964.
- ^ Hugenholtz P; et al. (1998). "Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity". Journal of Bacteriology. 180 (18): 4765–74. doi:10.1128/JB.180.18.4765-4774.1998. PMC 107498. PMID 9733676.
- ^ Marcy Y; et al. (2007). "Dissecting biological "dark matter" with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth". Proc Natl Acad Sci U S A. 104 (29): 11889–94. doi:10.1073/pnas.0704662104. PMC 1924555. PMID 17620602.
- ^ Wilson MC; et al. (2014). "An environmental bacterial taxon with a large and distinct metabolic repertoire" (PDF). Nature. 506 (7486): 58–62. Bibcode:2014Natur.506...58W. doi:10.1038/nature12959. PMID 24476823.
- ^ Dojka MA; et al. (2000). "Expanding the known diversity and environmental distribution of an uncultured phylogenetic division of bacteria". Applied and Environmental Microbiology. 66 (4): 1617–21. Bibcode:2000ApEnM..66.1617D. doi:10.1128/AEM.66.4.1617-1621.2000. PMC 92031. PMID 10742250.
- ^ Jon S. Graf, Sina Schorn, Katharina Kitzinger, Soeren Ahmerkamp, Christian Woehle, Bruno Huettel, Carsten J. Schubert, Marcel M. M. Kuypers & Jana Milucka: Anaerobic endosymbiont generates energy for ciliate host by denitrification. In: Nature (3 March, 2021). doi:10.1038/s41586-021-03297-6.
- ^ Mattila, J. T., N. Y. Burkhardt, H. J. Hutcheson, U. G. Munderloh, and T. J. Kurtti. 2007. Isolation of cell lines and a rickettsial endosymbiont from the soft tick Carios capensis (Neumann) (Acari: Argasidae: Ornithodorinae). Journal of Medical Entomology 44:1091–1101. "We propose that the isolate RCCE3 be named "Candidatus Rickettsia hoogstraalii" in honor of Harry Hoogstraal who contributed so much to our knowledge of ticks."
- ^ O. Mediannikov O, Aubadie-Ladrix M, Raoult D (2015). "Candidatus 'Rickettsia senegalensis' in cat fleas in Senegal". New Microbes New Infect. 3: 24–8. doi:10.1016/j.nmni.2014.10.005. PMC 4337942. PMID 25755888.
{{cite journal}}: CS1 maint: multiple names: authors list (link): "We isolated three rickettsial strains, and genetic analysis demonstrated that these strains represent a probable new species, provisionally called Candidatus Rickettsia senegalensis here." - ^ Tamaki H; et al. (2010). "Armatimonas rosea gen. nov., sp. nov., a Gram-negative, aerobic, chemoheterotrophic bacterium of a novel bacterial phylum, Armatimonadetes phyl. nov., formally called the candidate phylum OP10". International Journal of Systematic and Evolutionary Microbiology. 61 (Pt 6): 1442–7. doi:10.1099/ijs.0.025643-0. PMID 20622056.
- ^ Lee KCY; et al. (2010). "Chthonomonas calidirosea gen. nov., sp. nov., an aerobic, pigmented, thermophilic microorganism of a novel bacterial class, Chthonomonadetes classis. nov., of the newly described phylum Armatimonadetes originally designated candidate division OP10" (PDF). International Journal of Systematic and Evolutionary Microbiology. 61 (Pt 10): 2482–90. doi:10.1099/ijs.0.027235-0. PMID 21097641.
- ^ Mori K; et al. (2009). "Caldisericum exile gen. nov., sp. nov., an anaerobic, thermophilic, filamentous bacterium of a novel bacterial phylum, Caldiserica phyl. nov., originally called the candidate phylum OP5, and description of Caldisericaceae fam. nov., Caldisericales ord. nov. and Caldisericia classis nov". International Journal of Systematic and Evolutionary Microbiology. 59 (Pt 11): 2894–8. doi:10.1099/ijs.0.010033-0. PMID 19628600.
- ^ Geissinger O; et al. (2009). "The Ultramicrobacterium Elusimicrobium minutum gen. nov., sp. nov., the first cultivated representative of the Termite Group 1 Phylum". Applied and Environmental Microbiology. 75 (9): 2831–40. Bibcode:2009ApEnM..75.2831G. doi:10.1128/AEM.02697-08. PMC 2681718. PMID 19270135.
- ^ Podosokorskaya OA; et al. (2013). "Characterization of Melioribacter roseus gen. nov., sp. nov., a novel facultatively anaerobic thermophilic cellulolytic bacterium from the class Ignavibacteria, and a proposal of a novel bacterial phylum Ignavibacteriae". Environ. Microbiol. 15 (6): 1759–71. doi:10.1111/1462-2920.12067. PMID 23297868.
- ^ Cho JC; et al. (2004). "Lentisphaera araneosa gen. nov., sp. nov, a transparent exopolymer producing marine bacterium, and the description of a novel bacterial phylum, Lentisphaerae". Environmental Microbiology. 6 (6): 611–21. doi:10.1111/j.1462-2920.2004.00614.x. PMID 15142250.
- ^ Lucker S; et al. (2013). "The genome of Nitrospina gracilis illuminates the metabolism and evolution of the major marine nitrite oxidizer". Front. Microbiol. 4: 27. doi:10.3389/fmicb.2013.00027. PMC 3578206. PMID 23439773.








