Talk:DNA sequencing/Archive 1
| This is an archive of past discussions. Do not edit the contents of this page. If you wish to start a new discussion or revive an old one, please do so on the current talk page. |
| Archive 1 |
"Less tech" scratch-pad
Hmm, let's see:
- "DNA sequencing is any method that reveals any part of the central building block pattern that makes up a cell."
Evolve the article forward from there. Also, we are going to need a recognizable and clear history section. More illustrations too. Need to work in somethings about why we sequence DNA: being unique like fingerprints, find out how biology works, ... --Charles Gaudette 09:24, 18 July 2006 (UTC)
In near future, people's official personal name will be a babbled hash of their full DNA sequencing.
If and when it becomes possible to sequence the entire human DNA in short time (minutes) using small equipment (desktop or portable) then a hash algorythm could be developed, which can give a short unique identifier for any human on Earth, based on their full DNA seq - just like 512-byte little SHA-1 hashes are used to positively identify gigabyte sized files of digital data.
The hash number sequence could be converted to S-code (a vocabulary of artifical words) to make up an easy to remember sentence. This is already done in SSH Babble format for user friendly host key fingerprints. Similarly the DNA's spellable S-code hash would become your official name. I recommend using chinese names and/or native american names for the DNA's Babble dictionary. Even if you were John Doe Jr., your official genetic name will be something like "Pei Xio Li Bai Chung Zheng Kuo" or "Little Blue Cloud Sitting Wolf Three Arrows".
The results would be great. Personal identity cards would become unnecessary, as your DNA hash becomes your worldwide unique personal name and identity and it can be verified infallibly in situ from the blood or mouth swab using rapid desktop sequencers. Thus, it becomes impossible to live under false names. Murder victims can always be identified immediately. Murder suspects can be named immediately on national TV, even if they only left a drop of blood or spit behind.
Of course the hash forming algorithm needs to be designed carefully so that it is not possible to deduce private info e.g. about gender, racial identity or hereditary diseases just by knowing someone's hash. Such info shall be gained only from full sequencing.
I do not think this bright or menacing (see 666) future is very far. About 97% of human DNA is invariably same among all people so only the rest 3% needs to be sequnced and reduced to a unique hash. Sequencing technology is developing rapidly. 195.70.48.242 20:33, 5 December 2006 (UTC)
Samples with external dna often discarded after sequencing
We may need to rethink that that after this study. [1] Brian Pearson 01:32, 3 September 2007 (UTC)
new "Challenges" section
I feel this should simply be integrated into the section as a description of the resulting data, eliminate any attempts at comparison/contrast with the high-throughput methods.
- Paragraph 1: true, but this isn't a challenge, it's simply a description of the output.
- Paragraph 2: This isn't true, I sequence things all the time with a PCR primer, you only need to attach linkers for sequencing a library. In addition, you need to attach linkers for the high-throughput strategies too -- there's a PCR step, and there needs to be a common sequence for the sequencing primer. I believe in the traditional method you're putting them into cloning vectors to enable clonal isolation. As far as I understand it (and in my experience), filtering out these linker sequences is trivial, so I don't see how this information is notable.
- Paragraph 3: Most of the high throughput methods, with the notable exception of Helicos, also use PCR amplification; however, I think the problems described are probably more likely to arise in longer PCR products (high throughput library molecules are very short). Again, this is can be included as a description of the output.
Madeleine ✉ ✍ 13:45, 17 April 2008 (UTC)
improve intro
the current intro reads "The term DNA sequencing encompasses biochemical methods for determining the order of the nucleotide bases, adenine, guanine, cytosine, and thymine, in a DNA oligonucleotide. The sequence of DNA constitutes the heritable genetic information in nuclei, plasmids, mitochondria, and chloroplasts that forms the basis for the developmental programs of all living organisms." 1) why biochemical methods - seem overly restirictive 2) many organisms and viruses have nucleotides other then the canonical 4; even humans have a significant anount of 5 Methyl C 3) nuclei....chlorplast this could just be replaced by all organisms, with nod at RNA viruses 4)order of bases...I don't thinkk this is comprehensible to a layman; do we need a note about how dna is organized or a pictureCinnamon colbert (talk) 13:55, 13 June 2008 (UTC)
early methods
Add work from Ray Wu at cornell ? this is a little outside my knolwedge base - I don't know how important his work was. Cinnamon colbert (talk) 14:15, 20 April 2008 (UTC)
- role of sequenase - as someone who was sequencing, and wathcing others sequence, the transistion from maxam gilbert to sanger chain terminatin was slowed by the laborious protocol of sanger, eg using siliconized glass capillarys and klenof; the lazy americans, with their microfuge tubes, or even 96 well plates, plus S35 + sequenase really democritized sequencing Cinnamon colbert (talk) 12:37, 21 July 2008 (UTC)
Other "Next Generation" technologies?
I have added a lot of material here - if you have some comments, please let me knowCinnamon colbert 04:03, 29 March 2007 (UTC)
- I agree this article needs to be edited to make it more accessible and we should also add some links to the other "Next generation" technologies, e.g. Solexa and 454's biggest near-term competitor - Applied Biosystems' "SOLiD" platform - horrible name, nice technology! Also some discussion of the longer term technologies being investigated, such as nanopore sequencing? P.S. I don't work for Applied Biosystems!
- I tried reading the solid pdf, and found it awfully complex, a la the Brenner /Lynx paper - is this really goign to fly in the real world ??
- I agree this article needs to be edited to make it more accessible and we should also add some links to the other "Next generation" technologies, e.g. Solexa and 454's biggest near-term competitor - Applied Biosystems' "SOLiD" platform - horrible name, nice technology! Also some discussion of the longer term technologies being investigated, such as nanopore sequencing? P.S. I don't work for Applied Biosystems!
- request As I reviewed your next generation sequencing entries, I noticed you missed a technology that is currently commercialized = Solexa's next generation Sequencing By Synthesis platform. I'd like to request a link to our web page at solexa.com. Please let me know what I can provide to accomplish this. Glenn Powell Director, Marketing Solexa Inc. gpowell@solexa.com
- after looking at the illumina web site today (28 march 2007) I'm not sure I'd call the Solexa technolog "commercialized" in the sense that there are SKUs and pricing and so forth..looks a little custom at the momentCinnamon colbert 04:03, 29 March 2007 (UTC)
- I'd say that, as of today, both 454 and Solexa/Illumina can now be safely described as "commercialized". They've both shipped a more than decent numbers of platforms and you see data generated with them popping up about everywhere. Is there anything speaking against giving them a place in the "Major landmarks in DNA sequencing" section, like for the old Sanger-type ABIs 3xx and 3xxx? As to whether SOLiD should already appear there ... I'd still wait. BaChev (talk) 22:10, 26 August 2008 (UTC)
- after looking at the illumina web site today (28 march 2007) I'm not sure I'd call the Solexa technolog "commercialized" in the sense that there are SKUs and pricing and so forth..looks a little custom at the momentCinnamon colbert 04:03, 29 March 2007 (UTC)
- request As I reviewed your next generation sequencing entries, I noticed you missed a technology that is currently commercialized = Solexa's next generation Sequencing By Synthesis platform. I'd like to request a link to our web page at solexa.com. Please let me know what I can provide to accomplish this. Glenn Powell Director, Marketing Solexa Inc. gpowell@solexa.com
Since I’m close to the events described here, I’ll try to refrain from directly editing the article, but I can point out potential factual errors and peer-reviewed citations allowing other editors to decide.
- [9] Nat Biotechnol. 2003 Jun;21(6):673-8. "Multiplexed genotyping with sequence-tagged molecular inversion probes" is a lovely paper but is not even close to being the correct reference for “Solid, which combines .. an oligo ligation strategy[9]”
Instead, the reference below is relevant (also one of the first next-generation sequencing papers): Shendure, J, et al. (2005) Accurate Multiplex Polony Sequencing of an Evolved Bacterial Genome. Science 309(5741):1728-32.
- OK
- “colony-based technique (from Mitra and Church at Harvard)” doesn’t really fit under “other proposals” but fits earlier since it is directly relevant to the Solexa and AB methods, also it might be worth pointing out that polymerase colonies do not involve bacterial colonies as used in previous sequencing methods.
- fix it if you want
Mitra,RD, Shendure,J, Olejnik,J, Olejnik,EK, and Church,GM (2003) Fluorescent in situ Sequencing on Polymerase Colonies. Analyt. Biochem. 320:55-65
- “commercialized by Integrated Genetics,[17]” This was not the company that commercialized it; it was Genome Therapeutics and that reference [17] is not even close to the Church & Gilbert technology. Furthermore, to declare that it was not a commercial success, seems to merit reconsideration considering that it was responsible for the first commercial genome sequence in 1994 and related projects worth over $100M in value (http://www.nature.com/ng/wilma/v13n1.867861436.html ) and lead a few years later to other solid-phase, cyclic hybridization and imaging methods including the ‘next-generation’ methods cited above.
- Genomie therapuetics was a spin off of integrated genetics, to my memory; I meant to refer to the automated blot processor that IG/GT developed; commercial success is an undfined term, but I think it includes sales , on a repeat basis, to many other unrelated entitys, and it includes repeat on going revenue; a grant proposal is not, however large, commercial success in my mind.
- In the section on “Major landmarks in DNA sequencing” Some of these don’t seem so major or relevant to sequencing.
1992 “Haseltine is shown dye-terminator sequencing technology” “2003 Cold Spring Harbor sponsors GeneSweep, a sweepstakes on the number of human genes.”
- The statement “potential to produce 106-fold greater sequencing data” seems to cry out for an external reference. It would be safer to say 100-fold, but even for that it’s hard to find a peer-reviewed economic analysis. One such analysis is in the Science paper above.
- “**First chromosome physical maps published:” This seems to be misformatted (should be on a new line) and also questionable whether 4 examples of maps are needed since their not really (a) representative, (b) relevant to sequencing, (c) relevant to a broad audience.
--George Church 06:12, 27 May 2007 (UTC)
Wow - George Church here !!
fellow wikians - quite an honor to have Dr. Church, although why he feels he can't contibute is a bit of a mystery.. Cinnamon colbert 17:49, 9 June 2007 (UTC)
- It's not too surprising if you check his discussion page -- he's just trying to avoid any conflict of interest issues. I'm glad you're implementing his suggestions, I was hesitant to do so myself for COI reasons, so I put it off and hoped someone else would. -- Madeleine 18:08, 9 June 2007 (UTC)
New sequencing methods
I rewrote this section (and changed the title), trying to give more explanation of general tactics taken in pursuit of high-throughput sequencing. I'm posting here now because I noticed that someone with experience with Solexa got told not to edit this section. I need to make sure you know that I'm member of George Church's lab. As I mentioned earlier on this talk page, I was hesitant to edit the section. In the end, I did, because I wanted to improve it. I tried very hard to make this NPOV, using the first reference as a guide for what technologies are "well-known" and trying my best to be even-handed about it. Madeleine ✉ ✍ 23:40, 20 July 2007 (UTC)
(niave readers) User Frankatca has been the champion of a company called ZS genetics, which by its own admission, has yet to sequence any DNA. I am sure Frankatca, on his user page, can provide you with more information. Since ZS genetics has yet to do any actual sequencing, more then a mention of their name seems inappropriate at this time.
ZS Genetics has been accepted as the seventh firm in a juried competition for the Archon Xprize, i.e. 100 human genomes to be sequenced in 10 days at an ongoing cost of $10,000 or less/genome with fewer than 1 error per 100,000bp. Frankatca (talk) 01:12, 27 August 2008 (UTC)
After the genome-the Chronome
There has been much talk about how the human genome was complied,using scientific methods,and certainly,compiling the human genome was one of the most extensive research projects in history. Over the years,people had studied DNA,and had learned how portions of the DNA molecule functioned. They were dealing with bits and pieces of DNA,not the entire DNA molecule. That changed when the human genome was compiled,because now the entire DNA molecule had been mapped. Unfortunately,the human genome project did not list all the genetic expressions in the genome in chronological order. That's like writing a history book,and forgetting to put dates in it. Suppose you had a history book that had no dates. When was world war two? Was it in 1968? When did the first astronauts land on the moon? Was it in 1935? Obviously a history book without dates is not very useful,because it does not place historical events in a chronological framework. For an example of a chronological framework,look at a calendar,a calendar is a good example of a chronological framework. When they compiled the human genome,they did not list any of the genetic expressions in chronological order,they did not bother to link them to any kind of chronological framework,hence,the human genome is about as useful as a history book that doesn't have any dates in it. A few years ago,I wrote an article for a well-known science magazine,and the subject of my article was compiling the human chronome. I coined the term chronome,which means a chronological sequence of genetic expressions. The word chronome is based on the word 'chronological' and the word 'genome'. Unfortunately,the editor of that science magazine did not publish my article. I can briefly explain how genetic expressions take place over a period of time. The first genetic expression occurs during the pre-natal stage. In this stage of human development,genetic expression of DNA in the embryo's cells causes the anatomical features of the embryo and placenta to be formed. Most genetic expression probably occurs in the pre-natal period. After the baby is born,there is another period of genetic expression,when certain genes cause the baby's teeth to appear(dentition). At puberty,there is a robust phase of genetic expression,many genes express their genetic function during puberty,and the attributes of adulthood become more highly ramified in this phase of life. In males,the expression of certain genes cause the boy's beard to grow,in females,there is an increase in the size of the breasts,and menstruation begins. Later in life,females experience menopause,which is triggered by certain genes that are expressed when the woman is about 45 or 50 years old. Finally,in old age,there may be some expression of genes that cause senility,specifically,genes that cause Alzheimer's disease,et cetera. That's a summary of the chromological framework for the human chronome. Each genetic expression should be identified with a number,called a chronomic number. Low chronomic numbers identify expressions during youth,high chronomic numbers identify expressions during old age. The first chronomic number would be the number one,which would identify the very first gene that was expressed in the pre-natal phase,the gene that began the growth of the embryo. During the early stages of pregnancy,the embryo is neither male nor female,so during that phase,there is only one chronomic series. After sexual differentiation begins,there are two sets of chronomic numbers,or two chronomic series. One series is the male series,and the other series is the female series,they may be marked with the letters M for male,and F for female. For example,the chronomic number M7523 stands for the seven thousand,five hundred twenty-third expression in the male series. The chronomic number F8449 stands for the eight thousand,four hundred forty-ninth expression in the female series. The concept of the human chronome has the potential to become a research project in the future,perhaps employing thousands of scientists. It may take years to compile the human chronome from beginning to end,using scientific methods.Anthony Ratkov —Preceding unsigned comment added by 69.221.72.176 (talk) 07:50, 7 October 2008 (UTC)
- Is this a request to add this information the article? If so, please note that Wikipedia cannot accept original research. I suggest you publish your findings in a reputable (generally peer reviewed) journal first. -- MarcoTolo (talk) 14:42, 7 October 2008 (UTC)
- i think this is a typical out to lunch amateur - this is already a large and active area of research; but, even if it is not orig research, it doesn't fall in dna sequencing article - appropriate for article on gene expression or soemthing like that. Further, the idea that genes are expressed in an orderly fashion with time contradicts a lot of what we know, which is that gene expression is actually a little random, at least at the single cell level, and that it varys a lot with your particular genome and the enviroment (womb = combination of mothers DNA and food, bacteria, etc). For instance, if the embryo gets infected with a virus, that will certainly generate new gene expression patterns.Cinnamon colbert (talk) 14:00, 26 November 2008 (UTC)
Comment not useful
- Hi ArmyOfFluoride, I'm sorry to say that I don't find your comments very helpful. They're perfunctory and not at all specific, and you announce them by pointing to your affiliation with a "rather prestigious research university", as if to say that if you're unable to understand the intricacies of sequencing from this article, the rest of the world likely traipses around in utter darkness. Let's not forget that the last US president graduated from an ivy-league college, and by all accounts he seems to belong to those who give more weight to divine providence and "creative" myths than to scientific rationalization and analysis. So while those with a formal education may possess valuable skills, it doesn't immunize them against misconceptions or makes them inordinately smarter. As for the article, there are already some diagrams and sketches, covering some of the methods and principles, and from your comments it's unclear which specific aspects you would like to have covered more. So you're welcome to share your thoughts and knowledge to improve this entry, but I would like to urge you to do so without passing judgement on the content that is there, and by implying that its current shortcomings contribute to the cultural erosion you allude to. It's easy to point out wrongs, but difficult to do things right. Sorry to be snide here, but comments like this really get my goat. Malljaja (talk) 22:17, 19 March 2009 (UTC)
I will try to find a link to the Lewis Thomas of DNA sequencing. MBCF 01:45, 8 December 2006 (UTC)
Major landmarks?
There haven't been any major landmarks since 1998? Wouldn't high-throughput parallel sequencing count? Rees11 (talk) 16:59, 7 May 2009 (UTC)
- I agree that the the laser scanned gel (abi, amersham, licro, mol dynamics [missed any])was a major period, and the newer methods (illumina, 454, etc) represent a major landmakr, with perhaps pyrosequencing the startCinnamon colbert (talk) 12:58, 22 May 2009 (UTC)
Role of DNA isolation
I use to have a book that described the history of dna purification methods, from the late 1800s to the 1960s. As someone who started grad school in the 1980s, it struck me how high molecular weight DNA and RNA, as pure, protein free preps, took a long time to obtain. The point of this is that DNA sequencing was dependnet on pure DNA; the chromatographic methods for sequenicng DNA were probably avialable in the 50s - or maybe even earlier - but pure dna was not.Cinnamon colbert (talk) 12:58, 22 May 2009 (UTC)
Example: Dye-terminator sequencing
Can you please give some examples of dye molecules for Dye-terminator sequencing and how it works ? Which molecular reactions occur between dye and nucleic acid ? Thanks. --89.156.63.176 (talk) 12:30, 24 May 2009 (UTC)
WikiEthics, WikiPurpose, Etc.
So I am a bioinformatics tech about to enter a PhD program and I am wondering what my ethical responsibilities may or may not be in editing this article. I work with Solexa Sequencing techniques right now, which have some pretty novel and exciting possibilities that are well worth mentioning, but I am sort of tooting my own horn which may be received poorly. Also, along the lines of the "layman's terms" point, to what extent can/should the easily comprehensible descriptions be supplemented by more arcane material that might be useful to other researchers like myself. I know that when I, personally, search for 454 or Solexa on the web I go straight to the company's websites, which are not always clear or thorough. Can we offer a description that ascends through the high school level to the collegiate? and beyond? --WillJeck 01:48, 22 March 2007 (UTC)
- WillJeck, your first instinct was right. Promoting your own product would specifically violate Wikipedia rules against self-promotion and advertising. If we allowed Wikipedia to include self-promotion, people on the payroll of companies would overwhelm volunteers.
- The problem is that you can (and probably will) give undue emphasis to commercial companies -- if not to your own companies, to the commercial companies in general, as distinct from academic bjectively (although if someone from the company itself wrote the article, it would seem to violate Wikipedia rules on advertising and self-promotion). A link to the company web site would be legitimate too, if the site actually has useful information. But it would be a problem (and violate NPOV) if you choose commercial sites over non-commercial sites.
- Why, for example, don't you link to the Nobel website, [2] which has lots of educational material on DNA sequencing written on the level of an intelligent high school student? There's a huge amount of free information on DNA sequencing, written by teachers and academics. [3]
- I realize that the commercial companies have made important contributions and deserve a place in the article. But we have to strike a balance, and it's very easy for the commercial interests to take over. And of all the commercial inroads in Wikipedia, this article is much less of a problem than others. Nbauman 17:47, 24 March 2007 (UTC)
urgh, this guy doesn't work for a nextgen sequencing company he's a researcher (if I've understood correctly). The article is woefully out of date regarding nextgen sequencing such as Solexa/454 and SOLiD sequecing platforms. If we prevented all researchers from posting because "they might be boasting about their work" Wikipedia would lack a lot of useful information. 193.62.203.214 (talk) 14:31, 28 July 2008 (UTC)
- 100% agree! Part of the success of Wikipedia is because people like to promote their own POV. Another part of its success is that they are required to do that using NPOV. I'd suggest that you should add content first, and let the community judge it later. If useful information is added as part of a commercial, strip it out and use what you can! Especially in science, companies can't go too far, their claims will be checked and if they constantly promise the moon and only deliver cheese, their reputation gets damaged. --Dan|(talk) 09:44, 15 June 2009 (UTC)
You should read Wikipedia:Conflict of interest. If you don't work for Solexa I don't think you have a COI. If you do have a COI, you can still edit, you just have to be careful and follow some rules. Personally, I would love to have you add something to the Landmarks section about so-called "next gen" sequencing. Rees11 (talk) 15:40, 15 June 2009 (UTC)
DNA Sequencing and the Human Genome - Hype and Promise
Cinnamon colbert 01:08, 4 April 2007 (UTC) let me know what you think of this paragraph, currently self rated at start- status. Cinnamon colbert 01:08, 4 April 2007 (UTC) the point I am trying to make is that the human genome has not yet, really, been sequenced, and that there is a great deal of hype in the supposedly scientific press.
- Good point, but under Wikipedia rules you can't say it yourself, you have to find a source to attribute it to. I think the part about the unsequenced segments is the interesting part. I remember reading in Science about the unsequenced areas.
- It would also be good to find an example of hype to knock down. Most of what I've read about DNA sequencing has been fairly measured and restrained. In Science and the NEJM, anyway. Nbauman 02:12, 4 April 2007 (UTC)
- The points have some validity, but one needs to be a bit more grown up about the whole issue. It is now well known that the benefits and results of sequencing the human genome have been overblown by the media (especially when it was a "hot" topic), and also at least by some scientists seeking funding for this enterprise. In addition, ignoring for a moment that its content is lacking NPOV, I do not think that this section really belongs here. This article describes the principles of available DNA sequencing techniques, not sequencing of the human genome in particular. So if it is to be retained, it would need a new home, or else a section that deals with difficult-to-sequence regions such centromers or telomeres, repeats, etc, and possible approaches to sequencing those.Malljaja 09:06, 4 April 2007 (UTC)
thanks for the feedback, cinnamon colbert
Removal of the section
I removed this section without making comment here, my apologies. Cinnamon colbert posted to my talk page, I realize I should have said something here, so I'm pasting it here.Madeleine ✉ ✍ 15:06, 20 July 2007 (UTC)
- limitations current technology in dna seq page
- why did you delete this ? even by the std you cite, "complete = euchromatic" the "complete" chromosome seqs have gaps; I presume most of the gaps have been sized by southerns with pfge (which as you know was a schwartz cantor thing originally)
- I feel we are doing a dis service to the non prof community by failing to point out the tremendous gap between reality and hype.Cinnamon colbert 13:53, 20 July 2007 (UTC)
- As I understand it, the general issue is not that the DNA wasn't sequenced, the issue is that we can't figure out how to stitch it together due to repetitiveness. As such, I don't think this warrants its own section, it belongs within the sequence assembly section. It's deceptive to tell the reader "it's a lie! they didn't sequence all of it!" when the stuff that wasn't "sequenced" almost certainly was — it was simply too repetitive to be assembled in a linear fashion. Like a jigsaw where all the pieces are the same shape and same color. Rather than having a high-level section whose sole purpose is to "cry foul", I think this belongs as a more NPOV clarification within the section on "large-scale sequencing". To that end, I have added two sentences to that section (and a reference & link to the publicly available paper) — do these sentences work for you? Madeleine ✉ ✍ 15:06, 20 July 2007 (UTC)
- I concur with Madeleine--the heading Hype and promise always seemed out of place and somewhat detracted from the fact that this is an entry that is focused mainly on DNA sequencing technology. As per my above comment left shortly after this section was introduced, I think its content needs a new home, or, as MP just did, should be integrated in the technical description of limitations of current seq technologies. I haven't scoured Wikipedia for articles devoted to DNA sequence assembly & annotation, but I'd expect that it deserves its own entry. Malljaja 19:08, 20 July 2007 (UTC)
- I may be wrong, but actually I think there are issues with sections simply not being sequenced, as discussed here. It describes how, in addition to the difficulties of assembling repetitive sequences, some sections may be resistant to sequencing. Perhaps it doesn't need its own section (and certainly not one entitled "Hype and promise"), but surely it warrants a mention? Ribrob (talk) 15:01, 10 September 2009 (UTC)
- I concur with Madeleine--the heading Hype and promise always seemed out of place and somewhat detracted from the fact that this is an entry that is focused mainly on DNA sequencing technology. As per my above comment left shortly after this section was introduced, I think its content needs a new home, or, as MP just did, should be integrated in the technical description of limitations of current seq technologies. I haven't scoured Wikipedia for articles devoted to DNA sequence assembly & annotation, but I'd expect that it deserves its own entry. Malljaja 19:08, 20 July 2007 (UTC)
- As I understand it, the general issue is not that the DNA wasn't sequenced, the issue is that we can't figure out how to stitch it together due to repetitiveness. As such, I don't think this warrants its own section, it belongs within the sequence assembly section. It's deceptive to tell the reader "it's a lie! they didn't sequence all of it!" when the stuff that wasn't "sequenced" almost certainly was — it was simply too repetitive to be assembled in a linear fashion. Like a jigsaw where all the pieces are the same shape and same color. Rather than having a high-level section whose sole purpose is to "cry foul", I think this belongs as a more NPOV clarification within the section on "large-scale sequencing". To that end, I have added two sentences to that section (and a reference & link to the publicly available paper) — do these sentences work for you? Madeleine ✉ ✍ 15:06, 20 July 2007 (UTC)
Changing costs of DNA sequencing
How about a section focusing on the cost of DNA sequencing, and how it has changed? Suggest something along the lines of[1] [2] [3] [4]:
| Year | Cost per finished base |
|---|---|
| 1990 | $10 |
| 2003 | $0.05-0.06 |
| 2005 | $0.01 |
Mike Chelen (talk) 18:19, 20 May 2010 (UTC)
Split
I think Sanger and Maxam-Gilbert sequencing are given too much weight here, and would like them to go (return in the case of Sanger) to there own pages. Narayanese (talk) 19:22, 4 June 2010 (UTC)
article has gotten worse over the last year
I made a lot of contributions a couple of years ago, and the article has gotten worse since then . For instance, the part about solexa: Bridge started with Crhis adams and steve kron at the WIBR; moved to mosaic, which went under and sold the IP to manteia which became solexa which became illumina..... So mentioning solexa is kind of talking about oldhistory; it might be corret to talk about how the dominant player, illumina, uses chemistry developed at solexa... where is pacbio and iontorrent ? Wikipedia dna sequencing article, ripCinnamon colbert (talk) 01:42, 26 August 2010 (UTC)
Obvious contradiction
Re: History -says sanger method was rapidly replaced by maxam gilbert in one paragraph, and then in the next it states that the sanger method rapidly replaced the maxam gilbert. I don't know which is true (I strongly suspect the latter) but the contradiction is glaring.
Answer: the 1st was plus minus, it preceeded maxam gilbert, I don't remember if the second, dideoxy, came out ahead or at the same time as maxam gilbert. The really important point is that prior to plus minus, sequencing had been very very hard - one person could, under favorable conditions, do a few bases a year. Plus minus showed how a person could do hundreds or thousands a year; it really showed people that easy sequencing was possible. Cinnamon colbert (talk) 00:57, 27 October 2010 (UTC) At least, that is my memory - but I'm a little young, you would have to talk to someone who is now in thier late 60s to get an accurate read
=even worse
the article is even worse now. in the maxam gilbert section - endlabeling with potassium ?? total gibberishCinnamon colbert (talk) 23:09, 15 December 2010 (UTC) Also, alhtough Lynx did "sell" some systems, it wasn't really a success; you could say the automated church blot stuff from the compnay in waltham, ma (i forget who) was a 1st next gen
some comments 14 jan 2011
ref 35 is wrong, i think what they want is this http://www.genome.gov/27541189 not sure sanger replaced maxam gilbert due to less radioactivity; I did labeling,a nd P32 is such a big deal (it has a14 day half life, so in six months or so it is gone) I would say that dideoxy, particulary with teh sequenase kit (USB) and the pharmacia kit, was much easier (and lower energy S32 dNTPs gave thinner bands on film) the chain termination section should make a clearer distinction between 1st gen methods (radioactive label) and 2nd gen methods (fluorescence) In my mind, the big breakthru after sanger was 1st the USB sequenase kit, which really made sequecing something any idiot could do, and then then the ABI equipment. between ABI and next gen (lynx) methods were things like the church's genomic blotting sequencing, which was semi commercialized by a company in waltham MA, and probably some other things (i heard rumors that hitachi had an automated gel pourer) the illumina section is slighlty confusing, in that teh use of "ddNTP" doesn't clearly tell teh reader that these are reversible terminators Helicos - it is NOT bright fluorophores, it is $$ optics (high NA) and $$ stages and Big lasers the solid section is incomprehensible (not surpsing, since SoLiD is incomprehensible; I've read the technical lit many times, and still don't get it -and i know a lot about dna sequencing. I think their literature leaves out some key points
the future stuff should point out that seq by hyb has been around for a long time (i remember when th bulgarians first came to the us)
I think logically the emulsion process - a key insight - comes afer brenner; yu could say logically that emulsion is the simpler better form of brenners bead work...Cinnamon colbert (talk) 03:40, 18 January 2011 (UTC)
DNA nanoball section
I just created a DNA nanoball page and added a section about it here. This was a grad school medical genetics project for UBC, and I have no affiliation with Complete Genomics. Any feedback would be greatly appreciated. Cheers, Spence. Suspencewl (talk) 06:39, 25 February 2011 (UTC)
Hi, we would share a link to a collection of NGS software (e.g. aligners, assembler, etc.) at this page. Users can easily obtain an overview on the available tools for analyzing their data. I put the link first under "See also" but Johnuniq remove it with a statement "not appropriate for an external link to be in 'See also'; please explain how link helps on talk page".
- Could any of you help to find an appropriate place to share the link? There are already external links in "See also", "Genome Technology Access Center",and that's why I put another link there too.
- The reason to share this tool collection link is that I think it should help NGS users to quickly gain an overview on what are available to help them dealing with the exploding amount of NGS data. — Preceding unsigned comment added by Leon mei (talk • contribs) 08:50, 1 April 2011 (UTC)
- Your edit added the following link to DNA sequencing#See also:
- From the comment on my talk page, I see that I overlooked an earlier external link that was also in that section. I have removed that link (which was Genome Technology Access Center). External links should never be in the "See also" section (see WP:ALSO) because that section is for navigation: finding related articles within Wikipedia.
- If an external link is helpful for the article, there can be an "External links" section where such a link can be placed (see WP:EL for information on what is regarded as "helpful"). I see that this page has no "External links" section—that is probably because there have been numerous attempts to spam links on this page. I do not have time now to think about this any further, but the next step would be to read WP:EL and decide whether any criteria there would support the inclusion of an external link. If so, you might suggest that here, and wait to see if there is a response from other editors. There are probably a hundred or more links which could be added to this article—generally, links which merely list items such as software packages would not be considered helpful for a general reader. Is there a Wikipedia article on NGS? If so, and if it is directly relevant, that could be in "See also", and the external link on the NGS page. Johnuniq (talk) 09:47, 1 April 2011 (UTC)
Formulation/Wording of the page
I am ready to help improve this page. Tell me what needs to be improved, what you do not understand.
I still do not understand how the POSITION of the fluorescent base is determined !! Renebach (talk) 17:05, 9 June 2011 (UTC) Cheers, René -- the next comment doesn't help. Please only constructive comments. Renebach (talk) 17:05, 9 June 2011 (UTC)
Graph of plummeting sequencing costs
Hi, I just wanted to float the idea of adding this graph somewhere near the top (or at the top) of the article (and it's from an authoritative source). It is the perfect demonstration of the revolution that's been happening in the past three years. I think this revolution is one of the most important things to know about DNA sequencing and should be highlighted. Objections?
--Qwerty0 (talk) 13:59, 13 December 2011 (UTC)
what does raw sequence mean ?
this is not a silly question; "raw sequence" usually means relatively short (< 1,000 nt) long reads with error rates of 0.1 to >1% To convert this into "finished" usable sequence, you need to know a lot of stuff. Lets say the error rate increased over the period of the graph; this would mean that the slope of a finished seq graph would be less steep signed cinnamon colbert — Preceding unsigned comment added by 68.236.121.54 (talk) 20:14, 13 March 2012 (UTC)
Electrophoresis methods
I've noticed there's very little discussion about the electrophoresis methods that are used. I'm studying multiplexed capillary electrophoresis, and its main application is DNA sequencing. Yet I couldn't find a single reference to capillary electrophoresis in the article, much less multiplexed capillary electrophoresis. I think there should be an entire section on methods of detection, including any and all electrophoresis methods that are currently used to detect the DNA once it's labeled and everything.Jojojlj (talk) 18:50, 15 April 2012 (UTC)
Content forks
I will be splitting some material, such as that on chain-termination sequencing, into their own pages (in this case, Sanger sequencing). It would be helpful to this page to have a more general discussion of these methods and their history while leaving the details to the new pages, as is seen with the next-generation sequencing methods and their respective pages. Chain termination sequencing specifically was merged into this article a few years ago; the field of DNA sequencing has changed noticeably since that time. Please contact me on my talk page with any concerns. §everal⇒|Times 15:03, 5 September 2012 (UTC)
- Hello Several Times, I support your idea of paring back these two sections—the entry has become too unwieldy with the addition of next-gen methods. Keeping it tight here and including relevant info elsewhere is indeed the way to go. Malljaja (talk) 15:37, 5 September 2012 (UTC)
ZS genetics
User Frankatca has been doing blatant commercial pumping for a company called ZS Genetics. ZS may be the best thing since sliced bread, but right now they have yet to do any actual sequencing here - I think in the software field, this is called vaporware. Perhpas user Frankatca can restrict his commercial activitys to his user page. Cinnamon colbert (talk) 22:51, 11 August 2008 (UTC) IF, and when ZS shows that they can acutally sequence DNA, and that they can do so in some manner that offers some advantage, I am sure the community here on wiki will be glad to make space.
- As of 22 May 2009, as best as I can tell from the ZS Gentics web site, no actual sequencing has yet been done. Further, the technology looks suspiciously similar to a 15 or 20 year old proposal from Joel Sussman of Los Alamos. Cinnamon colbert (talk) 12:46, 22 May 2009 (UTC)
Dr. Michal Janitz, Max Planck Institute for Molecular Genetics, book: Next Generation Genome Sequencing: Towards Personalized Medicine, published by Wiley-Blackwell (Chapter 9): “Direct Sequencing by TEM of Z-Substituted DNA Molecules” by William R. Glover, III and Dr. W. Kelley Thomas, Hubbard Center for Genomics Studies, University of New Hampshire. Frankatca (talk) 18:40, 27 May 2010 (UTC)
Microscopy and Microanalysis; Cambridge University Press; October, 2012 issue: DNA Base Identification by Electron Microscopy; David C. Bell, a1, W. Kelley Thomas, a2, Katelyn M. Murtagh, a3, Cheryl A. Dionne, a3, Adam C. Graham, a4, Jobriah E. Anderson, a2 and William R. Glover, a3
a1 School of Engineering and Applied Sciences, Harvard University, Cambridge, MA 02138, USA
a2 Hubbard Center for Genome Studies, University of New Hampshire, Durham, NH 03824, USA
a3 ZS Genetics, North Reading, MA 01864, USA
a4 Center for Nanoscale Systems, Harvard University, Cambridge, MA 02138, USA
From the abstract: "...Dramatic improvements in genomic research will require accurate sequencing of long (>10,000 base-pairs), intact DNA molecules. Our approach directly visualizes the sequence of DNA molecules using electron microscopy. This report represents the first identification of DNA base pairs within intact DNA molecules by electron microscopy. By enzymatically incorporating modified bases, which contain atoms of increased atomic number, direct visualization and identification of individually labeled bases within a synthetic 3,272 base-pair DNA molecule and a 7,249 base-pair viral genome have been accomplished. This proof of principle is made possible by the use of a dUTP nucleotide, substituted with a single mercury atom attached to the nitrogenous base. One of these contrast-enhanced, heavy-atom-labeled bases is paired with each adenosine base in the template molecule and then built into a double-stranded DNA molecule by a template-directed DNA polymerase enzyme. This modification is small enough to allow very long molecules with labels at each A-U position. Image contrast is further enhanced by using annular dark-field scanning transmission electron microscopy (ADF-STEM). Further refinements to identify additional base types and more precisely determine the location of identified bases would allow full sequencing of long, intact DNA molecules, significantly improving the pace of complex genomic discoveries." Frankatca (talk) 14:12, 13 October 2012 (UTC) it is NOT yet sequencing; yhou are a long way from sequencing, as any knowledgeale person can tell — Preceding unsigned comment added by 24.91.51.31 (talk) 02:22, 18 April 2013 (UTC)
Missing Information
According to the article, it is not possible to sequence a DNA without killing it. Thus, if you sequence a sperm, then, it's gone. Is that true?--144.122.104.211 (talk) 13:14, 29 January 2013 (UTC)
- This is really a philosophical question more than anything. DNA molecules aren't really what most people would consider "alive" - they're just that, molecules. They may encode the information required to build a living organism but they cannot do it on their own. So DNA isn't really "killed" by sequencing, it's just used. It may even be copied numerous times in the process. So yes, if a sperm sample is used in a DNA sequencing reaction, it will be impossible to get the "original" sample back. That usually is not a problem since even a small volume of sperm contains more than enough DNA for sequencing. §everal⇒|Times 14:48, 29 January 2013 (UTC)
philosophical is right..you could say that a small, circular viral genome could be read by pac bio, and that the molecule, after reading, could be transfected into a cell....(i think - I'm a little hazy on the exact molecular details of the rolling circle and fluorophores) — Preceding unsigned comment added by 24.91.51.31 (talk) 02:25, 18 April 2013 (UTC)
- It may be philosophical but mine is not a philosophical question. Think that you are trying to select specific sperms by lookng at the DNA. For example you are eliminating the ones which have DNA that causes "tendency to disease X." So, at the end, you have only the "healthy" ones. But if you destroy the sperm while looking at the DNA in the sperm, you have nothing at the end. Because you destroyed the sperm for learning. You have the knowledge but don't have the sperm itself. Therefore, this is not a philosophical scenario. So, is it possible to conserve the DNA "unharmed" at the end of the "reading"? — Preceding unsigned comment added by 144.122.104.211 (talk) 21:48, 2 May 2013 (UTC)
- All DNA is copied DNA, whether it's in a human cell or a sperm cell or the result of amplifying DNA for a sequencing reaction. If you only had one sperm cell you could still easily use a process like PCR to provide as many copies of the DNA within the sperm as you needed. The "original" DNA would be intact. §everal⇒|Times 23:14, 2 May 2013 (UTC)
- Thank you for the answer Times. I think this information should be in the article.--144.122.104.211 (talk) 22:15, 29 May 2013 (UTC)
- This isn't really correct. Yes, there could theoretically be some future method that would allow us to copy the sperm's DNA without killing it, but not yet. Think what happens during PCR. You heat the DNA nearly to boiling to separate the strands. This will kill any sperm (and almost all other types of cells too). Even if it could survive that, you're also flooding the cell with chemicals like dNTP's and foreign enzymes. And then you have the problem of getting large DNA molecules out of the cell without disrupting it. We are not going to be able to do this any time soon.
- And current sequencing technologies are much worse. They have all of the same disruptive problems as PCR, and in addition, most of them explicitly rely on breaking the target DNA into millions of pieces.
- Qwerty0 (talk) 03:44, 30 May 2013 (UTC)
- All DNA is copied DNA, whether it's in a human cell or a sperm cell or the result of amplifying DNA for a sequencing reaction. If you only had one sperm cell you could still easily use a process like PCR to provide as many copies of the DNA within the sperm as you needed. The "original" DNA would be intact. §everal⇒|Times 23:14, 2 May 2013 (UTC)
- It may be philosophical but mine is not a philosophical question. Think that you are trying to select specific sperms by lookng at the DNA. For example you are eliminating the ones which have DNA that causes "tendency to disease X." So, at the end, you have only the "healthy" ones. But if you destroy the sperm while looking at the DNA in the sperm, you have nothing at the end. Because you destroyed the sperm for learning. You have the knowledge but don't have the sperm itself. Therefore, this is not a philosophical scenario. So, is it possible to conserve the DNA "unharmed" at the end of the "reading"? — Preceding unsigned comment added by 144.122.104.211 (talk) 21:48, 2 May 2013 (UTC)
Technical Problem?
When I start reading I wondered why you have to duplicate and cut the DNA to decode its molecular structure. I asked myself "Can't you just 'read' the molecule?" when I realized that this is the actual problem.
It would be nice to explain this as a general problem in the introduction to the methods developed especially for DNA molecules. 134.28.77.173 (talk) 12:16, 21 January 2014 (UTC)
re intro, delete word complete
the intro implies that we have complete seq for the human genome. This is NOT true. We don't have seq for the centromeres, and given the way in which genomes have been assembled from either short reads (illimina, solid, 454, helicos) or from longer (ABI capillary) reads from cloned BAC/PAC inserts, it is difficult to prove that regions have not been missed; indeed, recent PacBio data suggests that short reads miss a lot. so, please remove the word "complete" PS: people state that the centromeres aren't "interesting"; but how do you know if you havn't sequenced them ? — Preceding unsigned comment added by 24.91.49.238 (talk) 23:46, 22 June 2014 (UTC)
Accuracy of sequencing
Can somebody correct the 'accuracy' column in the table based on single-reads at a realistic read-length? Currently the table says 99.9999% for Pac-Bio 'consensus accuracy', something that would apply to any technology and hence is certainly incorrect/irrelevant. For Illumina, at a Phred 30, realistic for the read-length/read-quantity given in the table (ie >90% of bases read are >Phred30 typically on MiSeq/HiSeq, remaining 10% discarded), accuracy would be 99.9% not the 98% reported (that would be more like Phred 15). Pat Heslop-Harrison 10:54, 27 May 2015 (UTC) — Preceding unsigned comment added by Pathh (talk • contribs)
- Most of the numbers in this table need sources to truly consider them accurate. Consensus accuracy may not even serve as a relevant comparison here. I'll see if there's a better way to compare methods, but barring that, we'll need a recent review or two to provide some reliable sources. JHCaufield - talk - 17:20, 27 May 2015 (UTC)
Review
Proposal for future changes and general review
General suggestions for improvement for this page: To begin with I think this page is relatively well put together, but needs some refinement and addition to reflect the current advances in technology. The introductory section should be improved by the addition of a sentence the introduces the shift from Sanger to next generation sequencing (NGS). The section The four canonical bases should probably be removed.
Under Use of Sequencing the information is cursory and mostly lacking in citations. Most of these subsections could be vastly improved with an additional sentence explaining methodology and a proper citation (e.g. Under Evolutionary Biology emphasize the use of comparative genomics to evaluate the similarity and differences of organisms in order to determine their lineage). This could be cited using a review of phylogenetic studies or the manual for bootstrap.
Under History then Next Generation sequencing methods only the Roche 454 technology is mentioned. This should be replaced by a general explanation of the advent of NGS and what it has enabled.
Under Advanced methods and de novo sequencing Bridge PCR should be removed and the general techniques of NGS (and thus bridge PCR) put under Basic Methods.
I love the new addition of DNA Nanoball Sequencing. If somebody has the relevant information, the parameters of this method should be added into the table at the top of Next-generation sequencing methods.
The Sample Preparation section, citations are needed and it should be broken into two sections. One for DNA sample preparation and one for RNA sample preparation and subsequent cDNA conversion.
Finally, if there is to be a section on Read Trimming we should link it out to a main article discussing it if possible and a section should be added on read assembly and/or alignment
As a note of my personal background, I am a masters candidate specializing in NGS transcriptomic analysis. I look forward to feedback and hope some others will take part in the upgrading/updating of this page over the next few weeks. Krusec (talk) 02:25, 22 October 2015 (UTC)
- Hello All, in addition to Krusec's comments, it is a mammoth work but it is probably too technical for your average reader who simply came here to find out - in a nutshell - what is DNA sequencing. (Yes, how long is a piece of string.) However, it would benefit from one section near the beginning that is a very rough diagrammatic overview. I have cobbled together something from other DNA-related pages below but I am certainly no expert. Perhaps someone who is might tailor it, or something similar, for public consumption. We have the materials across Wikipedia; it is only a matter of assembling them. Regards, William Harris • talk • 20:40, 18 December 2015 (UTC)
-
The 33,000 year old skull of the "Altai dog"
-
Location of nuclear DNA within the chromosomes
-
The structure of part of a DNA double helix
-
An example of the results of automated chain-termination DNA sequencing
-
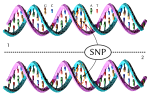
DNA molecule 1 differs from DNA molecule 2 at a single base-pair location - a single-nucleotide polymorphism
-
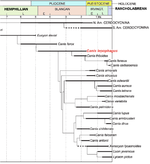
Phylogenetic tree and timeline towards the dog (Tedford 2009)
About the Sequence of the Pieces of DNA
If DNA is destroyed into millions of pieces before reading, how do they know which piece is after the other?--95.10.139.121 (talk) 17:53, 26 May 2015 (UTC)
- This is the problem of sequence assembly, either by mapping the pieces to an existing DNA sequence, or by assembling the fragments without any other information (de novo transcriptome assembly). --Amkilpatrick (talk) 19:00, 27 May 2015 (UTC).
Don't you need to sequence overlapping fragments ? (that's how I did it 1978). We were happy when we could read more than 100 bp per sequence. Renebach (talk) 13:06, 22 October 2016 (UTC)
much improved
from the last time I was here; at least, I don't think there are any really stupid errors (sad to say, there were) The whole next gen thing seems a little disorganized and repetitive; perhaps re organizing: by commercial, by date of intro, and non commercial, by date of first real sequence (say 100 bp or bases), and prospsective, no actual sequence data yet
if you talk to real sequencers, they will tell you that post sequence bio informatics is half of the work, so that section needs to be expanded - eg, a sequnce isn't any good if you don't know what to do with it — Preceding unsigned comment added by 50.245.17.105 (talk) 15:52, 31 December 2015 (UTC)
Added cPAS section to text and Table to describe technologies I work for BGI and MGI who build and sell DNA sequencers. I have added text with appropriate references to back up claims. I have not added any text to advantages or disadvantages columns in the Table so as not make any misleading comments. (AntoBeck (talk) 04:07, 5 July 2018 (UTC))
Whoa, where's the "layman's terms"?
Aww, I was hoping to learn how DNA was sequenced, but I reckon if you are an individual that understands "... initiated at a specific site on the template DNA by using a short oligonucleotide 'primer' complementary to the template at that region.", then you probably already know how DNA is sequenced. Common, there's got to be someone out there that is talented enough with words and biology to make this topic accessible to anyone who tries. :) -Tom
- I'll come back later and give it a try. I'll leave a note here so I'll remember to come back.Nbauman 19:15, 3 November 2006 (UTC)
This is quandry. Since Wikipedia is an encyclopedia it should be written for senior in high school level. But a senior should know what the four common DNA nucleotides are, that DNA has polarity, what a primer is, and that a primer annealing to a complementary strand could be used to initiate polymerization. But as I write this I can understand. To a molecular biologist DNA sequencing is as straight forward as it gets. To try to explain DNA sequencing and having to explain it all the way down to the basics of DNA polarity is a mind boggling task.
As a 3rd year molecular biology student at a rather prestigious research university I think I can say that DNA sequencing is not "as straight forward as it gets". This page could really use a diagram like the one on the PCR page. Sure, one might be well off to brush up on DNA Replication and PAGE before trying to tackle the intricacies of sequencing, but it might not hurt for the page to be a little clearer on why knowing the length of various segments tells one anything about which nucleotide goes where. As those in the know, we have a responsibility to share our knowledge with others, and I really think that its attitudes like this that have led whatever majority of americans to believe that the world was created in 6 days. So lets be helpful, if only for our own sake ArmyOfFluoride (talk) 05:16, 19 March 2009 (UTC) No such thing as DNA or not, doesn't matter. Be/can be any no matter what and any can be perfect. — Preceding unsigned comment added by VunslK (talk • contribs) 07:51, 28 September 2018 (UTC)
I feel like most methods under 'Methods in development' are included in the definition High-throughput methods
I propose to make Methods in dev under High-throughput methods for more clarity.Walidou47 (talk) 13:11, 11 February 2020 (UTC)
addition of new result by the NIH july 2020
Researchers have for the first time eve been able to sequence an entire human chromosome the x chromosome.
as some of you may know the human genome project never atually mapped the whole human genome so this result is quite important and exiting
im asking if this result can be added to the article and whare. — Preceding unsigned comment added by RJJ4y7 (talk • contribs) 18:49, 19 July 2020 (UTC)
Wiki Education Foundation-supported course assignment
![]() This article is or was the subject of a Wiki Education Foundation-supported course assignment. Further details are available on the course page. Student editor(s): Krusec. Peer reviewers: Krusec.
This article is or was the subject of a Wiki Education Foundation-supported course assignment. Further details are available on the course page. Student editor(s): Krusec. Peer reviewers: Krusec.
Above undated message substituted from Template:Dashboard.wikiedu.org assignment by PrimeBOT (talk) 18:57, 16 January 2022 (UTC)
Wiki Education Foundation-supported course assignment
![]() This article was the subject of a Wiki Education Foundation-supported course assignment, between 12 May 2020 and 22 June 2020. Further details are available on the course page. Peer reviewers: Cstaheli.
This article was the subject of a Wiki Education Foundation-supported course assignment, between 12 May 2020 and 22 June 2020. Further details are available on the course page. Peer reviewers: Cstaheli.
Above undated message substituted from Template:Dashboard.wikiedu.org assignment by PrimeBOT (talk) 19:45, 17 January 2022 (UTC)
Wiki Education Foundation-supported course assignment
![]() This article was the subject of a Wiki Education Foundation-supported course assignment, between 29 March 2021 and 4 June 2021. Further details are available on the course page. Student editor(s): Shady2021.
This article was the subject of a Wiki Education Foundation-supported course assignment, between 29 March 2021 and 4 June 2021. Further details are available on the course page. Student editor(s): Shady2021.
Above undated message substituted from Template:Dashboard.wikiedu.org assignment by PrimeBOT (talk) 19:45, 17 January 2022 (UTC)
this article is getting longer, fancier, but not better
I actually know something about DNA sequencing (I have done maxam gilbert, sanger, sequenase/sanger, minion, ilmn, pacbio) and the article is just a mess ah well — Preceding unsigned comment added by 50.245.17.105 (talk) 15:59, 14 March 2022 (UTC)
some history for you Noobs
back in 2007 or there bouts, a person called cinnamon colbert complained that the human genome hadn't actually been sequenced and this guy was mocked and jeered and reverted well, today Nature , one of the most prestiqous scientific journals in the entire world, says, quote seeems like you all owe colbert a big apology quote Fully finished genomes Roughly one-tenth of the human genome remained uncharted when genomics researchers Karen Miga at the University of California, Santa Cruz, and Adam Phillippy at the National Human Genome Research Institute in Bethesda, Maryland, launched the Telomere-to-Telomere (T2T) consortium in 2019. Now, that number has dropped to zero. In a preprint published in May last year, the consortium reported the first end-to-end sequence of the human genome, adding nearly 200 million new base pairs to the widely used human consensus genome sequence known as GRCh38, and writing the final chapter of the Human Genome Project1. — Preceding unsigned comment added by 50.245.17.105 (talk) 17:16, 4 February 2022 (UTC)
also, see the abstract to this article
https://www.biorxiv.org/content/10.1101/2022.06.24.497523v1
you people are so out of touch — Preceding unsigned comment added by 2601:192:4700:1F70:BCF8:D4F8:4FBA:FBA1 (talk) 13:09, 27 June 2022 (UTC)
table of robots in sample prep is spam for opentrons
yess the opentrons is much cheaper, but it is is much harder to program so you have to pay a lot of money for programming and it is very limited this table reads like an advert for opentrons and also this is wikipedia why do we need this table at all ?????? — Preceding unsigned comment added by 2601:192:4700:1F70:3C07:FF83:6CC9:713 (talk) 13:23, 27 June 2022 (UTC)